bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3134_orf1
Length=101
Score E
Sequences producing significant alignments: (Bits) Value
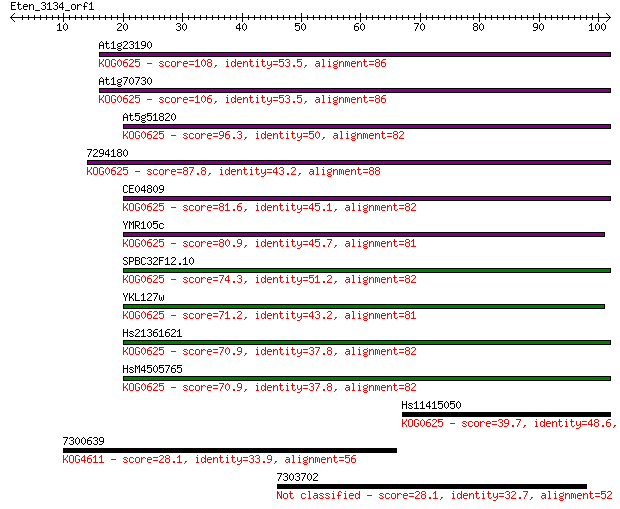
At1g23190 108 3e-24
At1g70730 106 1e-23
At5g51820 96.3 1e-20
7294180 87.8 4e-18
CE04809 81.6 3e-16
YMR105c 80.9 5e-16
SPBC32F12.10 74.3 5e-14
YKL127w 71.2 4e-13
Hs21361621 70.9 6e-13
HsM4505765 70.9 6e-13
Hs11415050 39.7 0.001
7300639 28.1 3.8
7303702 28.1 4.5
> At1g23190
Length=583
Score = 108 bits (269), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 65/86 (75%), Gaps = 0/86 (0%)
Query 16 SVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRY 75
S +T+P +GQKPGTSGLR+K + F QP YL N+VQA F+ L +V+G T++V+GDGRY
Sbjct 6 STVSTSPIDGQKPGTSGLRKKVKVFKQPNYLENFVQATFNALTAEKVKGATLVVSGDGRY 65
Query 76 FCSDAIKKIVAIAAGNGVGRVWIAKD 101
+ DA++ I+ +AA NGV RVW+ K+
Sbjct 66 YSKDAVQIIIKMAAANGVRRVWVGKN 91
> At1g70730
Length=585
Score = 106 bits (264), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 46/86 (53%), Positives = 65/86 (75%), Gaps = 0/86 (0%)
Query 16 SVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRY 75
S+ +T+P +GQKPGTSGLR+K + F QP YL N+VQA F+ L +V+G T++V+GDGRY
Sbjct 7 SLVSTSPIDGQKPGTSGLRKKVKVFKQPNYLENFVQATFNALTTEKVKGATLVVSGDGRY 66
Query 76 FCSDAIKKIVAIAAGNGVGRVWIAKD 101
+ AI+ IV +AA NGV RVW+ ++
Sbjct 67 YSEQAIQIIVKMAAANGVRRVWVGQN 92
> At5g51820
Length=623
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/82 (50%), Positives = 61/82 (74%), Gaps = 0/82 (0%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T P EGQK GTSGLR+K + FM+ YL+N++QA+F+ LP + + T+++ GDGRYF +
Sbjct 74 TKPIEGQKTGTSGLRKKVKVFMEDNYLANWIQALFNSLPLEDYKNATLVLGGDGRYFNKE 133
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
A + I+ IAAGNGVG++ + K+
Sbjct 134 ASQIIIKIAAGNGVGQILVGKE 155
> 7294180
Length=560
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 38/88 (43%), Positives = 61/88 (69%), Gaps = 1/88 (1%)
Query 14 SASVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDG 73
+ + AT P+EGQKPGTSGLR+K + F QP Y N+VQA+ + + + G T++V GDG
Sbjct 4 TVEIVATKPYEGQKPGTSGLRKKVKVFTQPNYTENFVQAILEA-NGAALAGSTLVVGGDG 62
Query 74 RYFCSDAIKKIVAIAAGNGVGRVWIAKD 101
R++C +A + IV ++A NGV ++ + ++
Sbjct 63 RFYCKEAAELIVRLSAANGVSKLLVGQN 90
> CE04809
Length=568
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 37/82 (45%), Positives = 53/82 (64%), Gaps = 0/82 (0%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T PF GQKPGTSGLR++ EF Q Y N+VQA+ D S+ +G ++V GDGR+ +
Sbjct 10 TKPFAGQKPGTSGLRKRVPEFQQQNYTENFVQAILDAGLGSKKKGSQLVVGGDGRFLSME 69
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
A I+ IAA NG+ R+ + ++
Sbjct 70 ATNVIIKIAAANGLSRLIVGQN 91
> YMR105c
Length=569
Score = 80.9 bits (198), Expect = 5e-16, Method: Composition-based stats.
Identities = 37/82 (45%), Positives = 56/82 (68%), Gaps = 2/82 (2%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T P+E QKPGTSGLR+KT+ F +P Y N++Q++ + +PE +G T++V GDGRY+
Sbjct 10 TKPYEDQKPGTSGLRKKTKVFKDEPNYTENFIQSIMEAIPEGS-KGATLVVGGDGRYYND 68
Query 79 DAIKKIVAIAAGNGVGRVWIAK 100
+ KI AI A NG+ ++ I +
Sbjct 69 VILHKIAAIGAANGIKKLVIGQ 90
> SPBC32F12.10
Length=554
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 42/82 (51%), Positives = 56/82 (68%), Gaps = 1/82 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSD 79
T P+EGQ+PGTSGLR+K F QP Y+ N+VQA D + E +G ++V GDGRYF
Sbjct 7 TKPYEGQRPGTSGLRKKVTVFEQPNYVENFVQATMDVV-EPSAKGAHLVVGGDGRYFNFH 65
Query 80 AIKKIVAIAAGNGVGRVWIAKD 101
AI+ I AIAAGNGV ++ + +
Sbjct 66 AIQVIAAIAAGNGVEKIIVGTN 87
> YKL127w
Length=570
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 52/82 (63%), Gaps = 2/82 (2%)
Query 20 TTPFEGQKPGTSGLRRKTREFM-QPGYLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T ++ QKPGTSGLR+KT+ FM +P Y N++QA +P G T++V GDGR++
Sbjct 10 TVAYKDQKPGTSGLRKKTKVFMDEPHYTENFIQATMQSIPNGS-EGTTLVVGGDGRFYND 68
Query 79 DAIKKIVAIAAGNGVGRVWIAK 100
+ KI A+ A NGV ++ I +
Sbjct 69 VIMNKIAAVGAANGVRKLVIGQ 90
> Hs21361621
Length=562
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPG-YLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T ++ QKPGTSGLR++ + F Y N++Q++ + ++ + T++V GDGR++
Sbjct 9 TQAYQDQKPGTSGLRKRVKVFQSSANYAENFIQSIISTVEPAQRQEATLVVGGDGRFYMK 68
Query 79 DAIKKIVAIAAGNGVGRVWIAKD 101
+AI+ I IAA NG+GR+ I ++
Sbjct 69 EAIQLIARIAAANGIGRLVIGQN 91
> HsM4505765
Length=562
Score = 70.9 bits (172), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 54/83 (65%), Gaps = 1/83 (1%)
Query 20 TTPFEGQKPGTSGLRRKTREFMQPG-YLSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCS 78
T ++ QKPGTSGLR++ + F Y N++Q++ + ++ + T++V GDGR++
Sbjct 9 TQAYQDQKPGTSGLRKRVKVFQSSANYAENFIQSIISTVEPAQRQEATLVVGGDGRFYMK 68
Query 79 DAIKKIVAIAAGNGVGRVWIAKD 101
+AI+ I IAA NG+GR+ I ++
Sbjct 69 EAIQLIARIAAANGIGRLVIGQN 91
> Hs11415050
Length=506
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 67 MLVAGDGRYFCSDAIKKIVAIAAGNGVGRVWIAKD 101
M+V DGRYF AI+ +V +AA NG+GR+ I ++
Sbjct 1 MVVGSDGRYFSRTAIEIVVQMAAANGIGRLIIGQN 35
> 7300639
Length=974
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 26/56 (46%), Gaps = 4/56 (7%)
Query 10 VPPESASVRATTPFEGQKPGTSGLRRKTREFMQPGYLSNYVQAVFDCLPESEVRGG 65
VP S + AT P + P T + T+ Y N +FDC+P +EV GG
Sbjct 23 VPICSLTTAATNPSQLPTPHTFRFQNPTQCLKNEFYNLN----LFDCVPCNEVAGG 74
> 7303702
Length=309
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 27/52 (51%), Gaps = 4/52 (7%)
Query 46 LSNYVQAVFDCLPESEVRGGTMLVAGDGRYFCSDAIKKIVAIAAGNGVGRVW 97
L Y +A+ + P + R T+ G +Y +D K+ V + GNG GR+W
Sbjct 179 LQRYTKAIAEA-PYKKQRNETL--GGFKKYLANDK-KQCVRVDQGNGYGRLW 226
Lambda K H
0.319 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184494980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40