bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3141_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
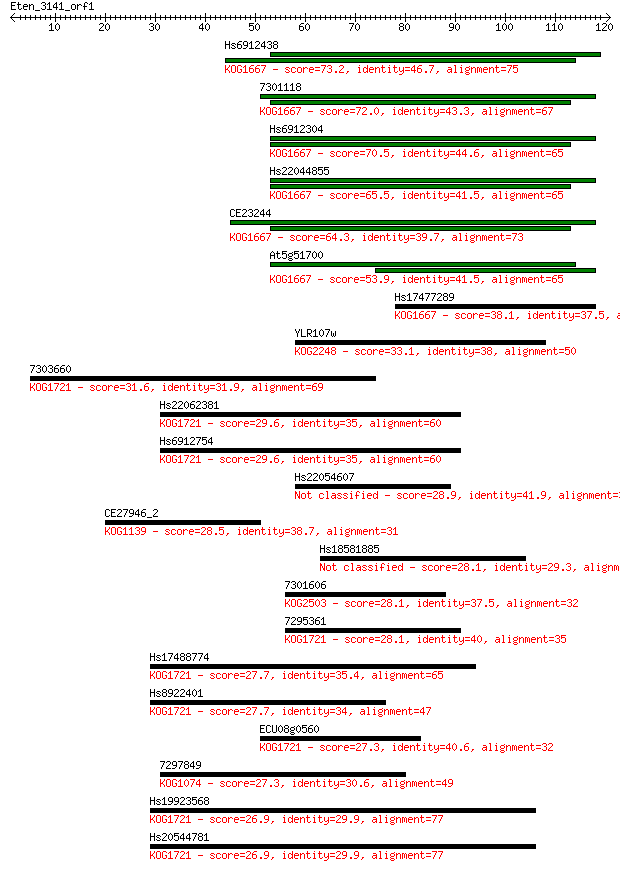
Hs6912438 73.2 1e-13
7301118 72.0 2e-13
Hs6912304 70.5 6e-13
Hs22044855 65.5 2e-11
CE23244 64.3 6e-11
At5g51700 53.9 7e-08
Hs17477289 38.1 0.004
YLR107w 33.1 0.14
7303660 31.6 0.34
Hs22062381 29.6 1.3
Hs6912754 29.6 1.4
Hs22054607 28.9 2.7
CE27946_2 28.5 3.1
Hs18581885 28.1 3.5
7301606 28.1 3.9
7295361 28.1 4.6
Hs17488774 27.7 5.3
Hs8922401 27.7 5.7
ECU08g0560 27.3 6.3
7297849 27.3 7.8
Hs19923568 26.9 8.0
Hs20544781 26.9 8.2
> Hs6912438
Length=347
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 32/67 (47%), Positives = 46/67 (68%), Gaps = 1/67 (1%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
CR GCGQ FDP++N DSC HH +P+FHD +K W CC + D+ F+ ++GC +GPH
Sbjct 5 CRNKGCGQHFDPNTNLPDSCCHHPGVPIFHDALKGWSCCRKRTVDFSEFLNIKGCTMGPH 64
Query 113 -SEEAPQ 118
+E+ P+
Sbjct 65 CAEKLPE 71
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 26/70 (37%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Query 44 MMAHGSSHKCRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMK 103
++ GSS C+ PGC + + C +H P FH+G+K W CCG + D+ AF+
Sbjct 142 LIRTGSS--CQNPGCDAVYQGPESDATPCTYHPGAPRFHEGMKSWSCCGIQTLDFGAFLA 199
Query 104 LQGCAVGPHS 113
GC VG H
Sbjct 200 QPGCRVGRHD 209
> 7301118
Length=354
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 29/67 (43%), Positives = 41/67 (61%), Gaps = 0/67 (0%)
Query 51 HKCRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVG 110
+C GCGQ FDP +N ++SCRHH P FHD K W CC +++ D+ F+ ++GC +
Sbjct 2 EQCYNRGCGQLFDPQTNNDESCRHHPGEPFFHDAYKGWSCCNKKSVDFTEFLNIKGCTLA 61
Query 111 PHSEEAP 117
HS P
Sbjct 62 KHSNVKP 68
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 0/60 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C+ GC +F +S+ C +H +P+FH+G+K W CC + D+ FM +GC G H
Sbjct 140 CKNNGCTYSFTGNSSDFGECTYHPGVPIFHEGMKFWSCCQKRTSDFSQFMAQKGCTYGEH 199
> Hs6912304
Length=332
Score = 70.5 bits (171), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C GCGQ FDP +N +D+C +H +PVFHD +K W CC + D+ F+ + GC G H
Sbjct 5 CYNRGCGQRFDPETNSDDACTYHPGVPVFHDALKGWSCCKRRTTDFSDFLSIVGCTKGRH 64
Query 113 SEEAP 117
+ E P
Sbjct 65 NSEKP 69
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 36/60 (60%), Gaps = 0/60 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C+ GC + + + E+ C +H +P+FH+G+K W CC ++ D++ F+ +GC G H
Sbjct 157 CKNGGCSKTYQGLESLEEVCVYHSGVPIFHEGMKYWSCCRRKTSDFNTFLAQEGCTKGKH 216
> Hs22044855
Length=259
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 27/65 (41%), Positives = 41/65 (63%), Gaps = 0/65 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C GCGQ FDP ++ +D+C +H +PVF+D +K W CC ++ D+ F+ + GC G H
Sbjct 5 CYNWGCGQRFDPETDSDDACTYHPGVPVFYDALKGWSCCKRKTTDFSDFLSIVGCTKGRH 64
Query 113 SEEAP 117
+ E P
Sbjct 65 NSEKP 69
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 35/60 (58%), Gaps = 0/60 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C+ GC + + + E+ C +H +P+FH+G+K CC ++ D++ F+ +GC G H
Sbjct 120 CKNRGCSKTYWGLESLEEVCVYHSGVPIFHEGMKYLSCCRRKTSDFNTFLAQEGCTTGKH 179
> CE23244
Length=321
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 29/73 (39%), Positives = 39/73 (53%), Gaps = 0/73 (0%)
Query 45 MAHGSSHKCRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKL 104
M S +C GCG FDP N ++C +H P FHD K W CC +++ D+ +M
Sbjct 1 MVDESKLQCYHKGCGLLFDPKENDNEACTYHPGGPYFHDAYKIWTCCDKKSTDFGTWMNY 60
Query 105 QGCAVGPHSEEAP 117
+GC G HS E P
Sbjct 61 KGCTRGKHSNEKP 73
Score = 57.0 bits (136), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 26/60 (43%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
CR GC FD S NKE+ C+HH +FH+G+K W CC ++ ++ AF++ GC G H
Sbjct 152 CRNNGCSTEFDGSKNKEN-CQHHPGAAIFHEGMKYWSCCNKKTSNFGAFLEQVGCTSGEH 210
> At5g51700
Length=208
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 25/61 (40%), Positives = 33/61 (54%), Gaps = 0/61 (0%)
Query 53 CRRPGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPH 112
C+ GCGQ F N E +C HH VFHD ++ W CC ++D FM++ C G H
Sbjct 141 CKNKGCGQTFKERDNHETACSHHPGPAVFHDRLRGWKCCDVHVKEFDEFMEIPPCTKGWH 200
Query 113 S 113
S
Sbjct 201 S 201
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 28/44 (63%), Gaps = 0/44 (0%)
Query 74 HHKLMPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPHSEEAP 117
+ L P FHDG+K+W CC Q + D+ F+++ GC G H+ E P
Sbjct 15 YSNLQPFFHDGMKEWSCCKQRSHDFSLFLEIPGCKTGKHTTEKP 58
> Hs17477289
Length=382
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 78 MPVFHDGVKQWPCCGQEAWDWDAFMKLQGCAVGPHSEEAP 117
+PVFHD +K W C + D+ F+ + C G H+ E P
Sbjct 240 VPVFHDALKGWSCYKRRTTDFSDFLSIVHCTKGRHNSEKP 279
> YLR107w
Length=404
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 19/53 (35%), Positives = 29/53 (54%), Gaps = 5/53 (9%)
Query 58 CGQAFDPSSNKEDS-CRHHKLMPVFHDGVK--QWPCCGQEAWDWDAFMKLQGC 107
C F + E + CR+H L +++ K Q+PCCG E D +F++L GC
Sbjct 159 CSSNFKKTDIMEKTLCRYHPLKRIYNRDTKNHQYPCCG-ETTDSVSFLRL-GC 209
> 7303660
Length=2532
Score = 31.6 bits (70), Expect = 0.34, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 28/73 (38%), Gaps = 4/73 (5%)
Query 5 PASSAESAPRVAAAPCSAARAAPCFPCCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQ 60
P + + P AAAP A + + C C C + H +H RP CG
Sbjct 415 PTTVSPGIPGGAAAPNPACEKSGRYVCQYCNLICAKPSVLEKHIRAHTNERPYPCDTCGI 474
Query 61 AFDPSSNKEDSCR 73
AF SN CR
Sbjct 475 AFKTKSNLYKHCR 487
> Hs22062381
Length=509
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 26/64 (40%), Gaps = 11/64 (17%)
Query 31 CCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHHKLMPVFHDGVK 86
C C C +M H SH RP CG +F + K + RH KL H G K
Sbjct 120 CDVCGLSCISFNVLMVHKRSHTGERPFQCNQCGASF---TQKGNLLRHIKL----HTGEK 172
Query 87 QWPC 90
+ C
Sbjct 173 PFKC 176
> Hs6912754
Length=509
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 26/64 (40%), Gaps = 11/64 (17%)
Query 31 CCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHHKLMPVFHDGVK 86
C C C +M H SH RP CG +F + K + RH KL H G K
Sbjct 120 CDVCGLSCISFNVLMVHKRSHTGERPFQCNQCGASF---TQKGNLLRHIKL----HTGEK 172
Query 87 QWPC 90
+ C
Sbjct 173 PFKC 176
> Hs22054607
Length=170
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 58 CGQAFDPSSNKEDSCRHHKLMPVFHDGVKQW 88
CGQ F PS +K D+ R+ + +P GV+ W
Sbjct 74 CGQLFKPSDDKPDNTRNERQIPE-ERGVQFW 103
> CE27946_2
Length=1919
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 15/34 (44%), Gaps = 3/34 (8%)
Query 20 CSAARAAPCFPCCCCCCCCCCC---CCMMAHGSS 50
C+ + P +P C C C CC MAH S
Sbjct 1775 CNKCSSVPSYPVMCLFCGEILCLNDCCRMAHQES 1808
> Hs18581885
Length=397
Score = 28.1 bits (61), Expect = 3.5, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 20/41 (48%), Gaps = 0/41 (0%)
Query 63 DPSSNKEDSCRHHKLMPVFHDGVKQWPCCGQEAWDWDAFMK 103
DP S+ E ++ P+F ++ C G+E WD M+
Sbjct 281 DPDSSLEGFALPMQMFPMFRKNFEKLECLGRERGQWDQNMQ 321
> 7301606
Length=1478
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 17/32 (53%), Gaps = 0/32 (0%)
Query 56 PGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQ 87
PG +F+P+ ED HH+ + HD V Q
Sbjct 685 PGPDPSFNPNIFSEDEDEHHQHQGIHHDAVPQ 716
> 7295361
Length=863
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 6/35 (17%)
Query 56 PGCGQAFDPSSNKEDSCRHHKLMPVFHDGVKQWPC 90
PGCGQ F+ N + RH+K++ H+ VK + C
Sbjct 639 PGCGQGFNNIGNMK---RHYKII---HEKVKDFAC 667
> Hs17488774
Length=568
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 23/71 (32%), Positives = 30/71 (42%), Gaps = 13/71 (18%)
Query 29 FPCCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHHKLMPVFHDG 84
FPC C C + H + H +RP CG AF S RH ++ H G
Sbjct 420 FPCPTCGKCFTKSSNLSEHQTLHTGQRPFKCADCGVAFAQPSRL---VRHQRI----HTG 472
Query 85 VKQWPC--CGQ 93
+ +PC CGQ
Sbjct 473 ERPFPCTQCGQ 483
> Hs8922401
Length=481
Score = 27.7 bits (60), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 22/51 (43%), Gaps = 4/51 (7%)
Query 29 FPCCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHH 75
+PC C ++ H +H RP CG+AF SSN + R H
Sbjct 204 YPCPQCGKAFGQSSALLQHQRTHTAERPYRCPHCGKAFGQSSNLQHHLRIH 254
> ECU08g0560
Length=449
Score = 27.3 bits (59), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 15/32 (46%), Gaps = 0/32 (0%)
Query 51 HKCRRPGCGQAFDPSSNKEDSCRHHKLMPVFH 82
HKCR PGC +AF S N + H H
Sbjct 403 HKCRFPGCSKAFSRSDNLSQHYKVHSTTNEMH 434
> 7297849
Length=1373
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 20/53 (37%), Gaps = 4/53 (7%)
Query 31 CCCCCCCCCCCCCMMAHGSSHKCRRPG----CGQAFDPSSNKEDSCRHHKLMP 79
C C C + H +H RP CG+AF N + HK+ P
Sbjct 826 CVVCDRVLSCKSALQMHYRTHTGERPFKCRICGRAFTTKGNLKTHMAVHKIRP 878
> Hs19923568
Length=595
Score = 26.9 bits (58), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 36/83 (43%), Gaps = 14/83 (16%)
Query 29 FPCCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHHKLMPVFHDG 84
+ C C C ++AH +H RP CG+AF S+ ++ R+ H G
Sbjct 306 YKCDQCGKAYGRSCHLIAHKRTHTGERPYECHDCGKAFQHPSHLKEHVRN-------HTG 358
Query 85 VKQWPC--CGQEAWDWDAFMKLQ 105
K + C CG+ A+ W + L
Sbjct 359 EKPYACTQCGK-AFRWKSNFNLH 380
> Hs20544781
Length=595
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 36/83 (43%), Gaps = 14/83 (16%)
Query 29 FPCCCCCCCCCCCCCMMAHGSSHKCRRP----GCGQAFDPSSNKEDSCRHHKLMPVFHDG 84
+ C C C ++AH +H RP CG+AF S+ ++ R+ H G
Sbjct 306 YKCDQCGKAYGRSCHLIAHKRTHTGERPYECHDCGKAFQHPSHLKEHVRN-------HTG 358
Query 85 VKQWPC--CGQEAWDWDAFMKLQ 105
K + C CG+ A+ W + L
Sbjct 359 EKPYACTQCGK-AFRWKSNFNLH 380
Lambda K H
0.327 0.135 0.532
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40