bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
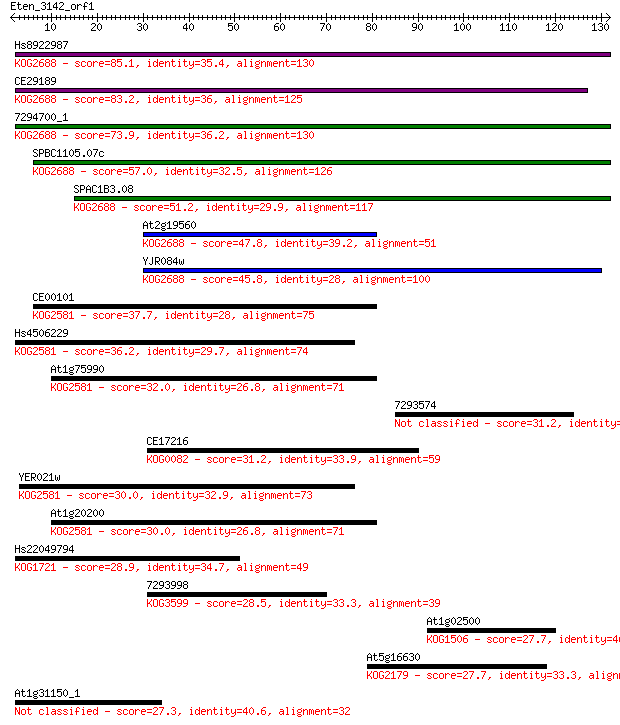
Query= Eten_3142_orf1
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
Hs8922987 85.1 3e-17
CE29189 83.2 1e-16
7294700_1 73.9 8e-14
SPBC1105.07c 57.0 1e-08
SPAC1B3.08 51.2 5e-07
At2g19560 47.8 5e-06
YJR084w 45.8 2e-05
CE00101 37.7 0.006
Hs4506229 36.2 0.015
At1g75990 32.0 0.31
7293574 31.2 0.52
CE17216 31.2 0.57
YER021w 30.0 1.1
At1g20200 30.0 1.2
Hs22049794 28.9 2.6
7293998 28.5 3.2
At1g02500 27.7 5.3
At5g16630 27.7 5.8
At1g31150_1 27.3 6.8
> Hs8922987
Length=453
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 46/130 (35%), Positives = 75/130 (57%), Gaps = 7/130 (5%)
Query 2 ILECLVAARLRMGKLPSADLLNKYQMPHYLDIIKAMKSGNLSLFDRALEQHESVFIKHGT 61
IL L+ ++ +G +P+ +LL KY + + ++ +A+ GNL L AL +HE+ FI+ G
Sbjct 305 ILIYLLPVKMLLGHMPTVELLKKYHLMQFAEVTRAVSEGNLLLLHEALAKHEAFFIRCGI 364
Query 62 ILCVERLKFIVFRTLMKRMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACI 121
L +E+LK I +R L K++ L K + + + F A+K+ E D DE+ CI
Sbjct 365 FLTLEKLKIITYRNLFKKVY-------LLLKTHQLSLDAFLVALKFMQVEDVDIDEVQCI 417
Query 122 SANLIRMGYI 131
ANLI MG++
Sbjct 418 LANLIYMGHV 427
> CE29189
Length=413
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/125 (36%), Positives = 74/125 (59%), Gaps = 7/125 (5%)
Query 2 ILECLVAARLRMGKLPSADLLNKYQMPHYLDIIKAMKSGNLSLFDRALEQHESVFIKHGT 61
IL L+ ++ +G +P++ LL++Y++ + D++ +K GNL+ D AL +E+ FIK G
Sbjct 263 ILIYLIPVKMFLGHMPTSQLLHEYRLDEFQDVVAGVKDGNLAQLDGALAANEAFFIKCGI 322
Query 62 ILCVERLKFIVFRTLMKRMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACI 121
L +E+L+ I FRTL K++ Q + A I P+ F A+++ D DE+ CI
Sbjct 323 FLMLEKLRMITFRTLFKKVSQ------IVGTAQI-PLDAFQTALRFVGVTDVDMDELECI 375
Query 122 SANLI 126
ANLI
Sbjct 376 IANLI 380
> 7294700_1
Length=372
Score = 73.9 bits (180), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 47/130 (36%), Positives = 72/130 (55%), Gaps = 7/130 (5%)
Query 2 ILECLVAARLRMGKLPSADLLNKYQMPHYLDIIKAMKSGNLSLFDRALEQHESVFIKHGT 61
IL LV ++ +G LPS LL +Y + +LD+ AMK+GN++ FD + E V I+ G
Sbjct 249 ILIYLVPVKMLLGYLPSKSLLQRYDLLLFLDLAMAMKAGNVNRFDEIVRDQELVLIRSGI 308
Query 62 ILCVERLKFIVFRTLMKRMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACI 121
L VE+LKF+V+R L K++ + K++ + + F +A+ + DE CI
Sbjct 309 YLLVEKLKFLVYRNLFKKV-------FVIRKSHQLDMGDFLSALHFVGLTDVSLDETHCI 361
Query 122 SANLIRMGYI 131
ANLI G I
Sbjct 362 VANLIYDGKI 371
> SPBC1105.07c
Length=442
Score = 57.0 bits (136), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 70/130 (53%), Gaps = 5/130 (3%)
Query 6 LVAARLRMGKLPSADLLNKYQMPHYLD-IIKAMKSGNLSLFDRALEQHESV--FIKHGTI 62
L L +GK P+ LL KY++ + +IKA+KSG++ F +LE + FIK G
Sbjct 269 LTTCLLILGKSPTKGLLEKYKLTAAFEPLIKALKSGDIKSFRLSLEDNSRRKWFIKRGIY 328
Query 63 LCV-ERLKFIVFRTLMKRMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACI 121
L + +R + I++R L +++ + E + ++ + TAA +D +D D++ CI
Sbjct 329 LTLLDRCEIILWRNLFRKVFLFTFEQSQKT-PHVSGSYLLTAARLSTNDNSYDMDDVECI 387
Query 122 SANLIRMGYI 131
+LI GYI
Sbjct 388 CVSLIDQGYI 397
> SPAC1B3.08
Length=423
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 66/121 (54%), Gaps = 13/121 (10%)
Query 15 KLPSADLLNKYQMPH----YLDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCVERLKF 70
+LP+ +LL+K+ P+ Y+ + +A+KSGNL F + L+++E++ K L +E +
Sbjct 287 QLPTKNLLSKF--PNLASVYIPLTRALKSGNLGEFGKCLQKNETLLAKTKIYLTLEGTRD 344
Query 71 IVFRTLMKRMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACISANLIRMGY 130
+ R L ++ W+ G K+ +P+ VF A++ +L + I AN+I GY
Sbjct 345 LCIRNLFRKT---WIICG---KSTRLPVSVFQIALQVAGTDL-PKLHVEAILANMISKGY 397
Query 131 I 131
+
Sbjct 398 M 398
> At2g19560
Length=244
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 35/51 (68%), Gaps = 0/51 (0%)
Query 30 YLDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCVERLKFIVFRTLMKRM 80
Y I++A++ G+L L AL++HE F++ G L +E+L+ V++ LMK+M
Sbjct 194 YTKIVQALRKGDLRLLRHALQEHEDRFLRSGVYLVLEKLELQVYQRLMKKM 244
> YJR084w
Length=423
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/104 (26%), Positives = 53/104 (50%), Gaps = 9/104 (8%)
Query 30 YLDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCVERLKFIVFRTLMKRMKQWWLESGL 89
Y +++++ SGNLSL++ HE F+ G + + L+ +VF L++R QW
Sbjct 300 YPTLVRSVISGNLSLYEATAASHERFFLSQGLHVVITLLREVVFTRLVQRCWQWG----- 354
Query 90 ASKANIVPIQVFTAAIKWQSDELFDDDE----MACISANLIRMG 129
+ +I+P+++ A + S D++E + C A+ I G
Sbjct 355 NDRKSIMPLKILLATKQHDSSANEDEEEQLDALECRLASAIASG 398
> CE00101
Length=504
Score = 37.7 bits (86), Expect = 0.006, Method: Composition-based stats.
Identities = 21/79 (26%), Positives = 45/79 (56%), Gaps = 4/79 (5%)
Query 6 LVAARLRMGKLPSADLLNK--YQ--MPHYLDIIKAMKSGNLSLFDRALEQHESVFIKHGT 61
+V L G++P + + Y+ + HYLD+ + ++ G+++ F+ LEQ ++ F T
Sbjct 300 VVVIGLLQGEIPDRSVFRQPIYRKCLAHYLDLSRGVRDGDVARFNHNLEQFKTQFEADDT 359
Query 62 ILCVERLKFIVFRTLMKRM 80
+ + RL+ V +T +K++
Sbjct 360 LTLIVRLRQNVIKTAIKQI 378
> Hs4506229
Length=534
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 43/79 (54%), Gaps = 6/79 (7%)
Query 2 ILECLVAARLRMGKLPS-----ADLLNKYQMPHYLDIIKAMKSGNLSLFDRALEQHESVF 56
+ + L+ L +G++P L + MP++L + +A+++GNL+ F++ L+Q F
Sbjct 328 VHKLLIVVELLLGEIPDRLQFRQPSLKRSLMPYFL-LTQAVRTGNLAKFNQVLDQFGEKF 386
Query 57 IKHGTILCVERLKFIVFRT 75
GT + RL+ V +T
Sbjct 387 QADGTYTLIIRLRHNVIKT 405
> At1g75990
Length=487
Score = 32.0 bits (71), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query 10 RLRMGKLPSADLLNKYQMPH----YLDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCV 65
RL +G++P + + M Y ++ A++ G+L LF + E+ F + T +
Sbjct 288 RLLLGEIPERSIFTQKGMEKTLRPYFELTNAVRIGDLELFGKIQEKFAKTFAEDRTHNLI 347
Query 66 ERLKFIVFRTLMKRM 80
RL+ V RT ++ +
Sbjct 348 VRLRHNVIRTGLRNI 362
> 7293574
Length=582
Score = 31.2 bits (69), Expect = 0.52, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Query 85 LESGLASKANIVPIQVFTAAIKWQSDELFDDDEMACISA 123
L +G+A A + V +KW E FDD+E+ CISA
Sbjct 112 LLAGVALNATVG--SVLLQPVKWHMKEEFDDEELMCISA 148
> CE17216
Length=329
Score = 31.2 bits (69), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 29/59 (49%), Gaps = 0/59 (0%)
Query 31 LDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCVERLKFIVFRTLMKRMKQWWLESGL 89
L I+KAM+ N+S D A E+ +FI H + L L MKQ W++ G+
Sbjct 81 LAILKAMQPLNISFTDAAREEDARMFISHFLHVNNAELSEAFSLELSDLMKQLWMDEGV 139
> YER021w
Length=523
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 41/79 (51%), Gaps = 10/79 (12%)
Query 3 LECLVAARLRMGKLPSADLLNKYQM-----PHYLDIIKAMKSGNLSLFDRALEQHESVFI 57
L C + +L MG +P ++ M P+Y + KA+K G+L F + +++ + +
Sbjct 316 LHCCI--QLLMGDIPELSFFHQSNMQKSLLPYY-HLTKAVKLGDLKKFTSTITKYKQLLL 372
Query 58 KHGTI-LCVERLKFIVFRT 75
K T LCV RL+ V +T
Sbjct 373 KDDTYQLCV-RLRSNVIKT 390
> At1g20200
Length=488
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 36/75 (48%), Gaps = 4/75 (5%)
Query 10 RLRMGKLPSADLLNKYQMPH----YLDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCV 65
RL +G++P + + M Y ++ A++ G+L LF E+ F + T +
Sbjct 289 RLLLGEIPERSIFTQKGMEKALRPYFELTNAVRIGDLELFRTVQEKFLDTFAQDRTHNLI 348
Query 66 ERLKFIVFRTLMKRM 80
RL+ V RT ++ +
Sbjct 349 VRLRHNVIRTGLRNI 363
> Hs22049794
Length=422
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 1/49 (2%)
Query 2 ILECLVAARLRMGKLPSADLLNKYQMPHYLDIIKAMKSGNLSLFDRALE 50
+L+ + RL + LP D+L H DI+K K G L DR+L+
Sbjct 79 LLDFMYEGRLDLRSLPVEDVLAAASYLHMYDIVKVCK-GRLQEKDRSLD 126
> 7293998
Length=728
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 0/39 (0%)
Query 31 LDIIKAMKSGNLSLFDRALEQHESVFIKHGTILCVERLK 69
L II+ G F R L H+ +FI G ++C+ L+
Sbjct 432 LQIIETATKGQYLDFQRPLRLHQMLFIVKGFLVCITTLR 470
> At1g02500
Length=393
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 92 KANIVPIQVFTAAIKWQSDELFDDDEMA 119
K +VPI+V T I Q DE +DE+A
Sbjct 180 KGAMVPIRVHTVLISTQHDETVTNDEIA 207
> At5g16630
Length=856
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 79 RMKQWWLESGLASKANIVPIQVFTAAIKWQSDELFDDDE 117
+ K+ WL GL KAN VP ++ K++ + F+D +
Sbjct 588 KTKERWLRDGLQLKANEVPSKILKRNSKFKKVKDFEDGD 626
> At1g31150_1
Length=551
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 2 ILECLVAARLRMGKLPSADLLNKYQMPHYLDI 33
I E + LR GKLP+ D + K+Q YL +
Sbjct 104 IREFHIVTGLRCGKLPTEDEVKKHQDSKYLSV 135
Lambda K H
0.327 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1283105810
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40