bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3156_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
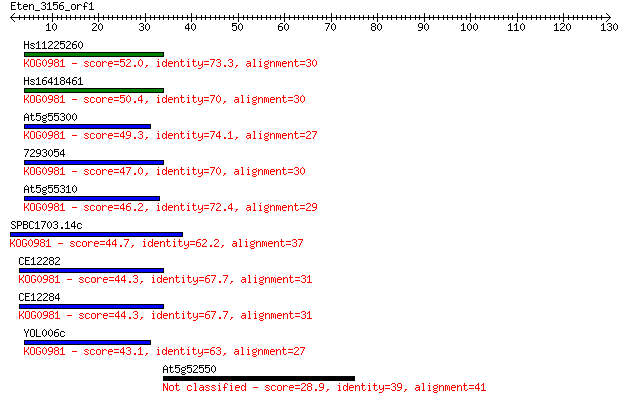
Hs11225260 52.0 2e-07
Hs16418461 50.4 8e-07
At5g55300 49.3 2e-06
7293054 47.0 9e-06
At5g55310 46.2 1e-05
SPBC1703.14c 44.7 4e-05
CE12282 44.3 5e-05
CE12284 44.3 6e-05
YOL006c 43.1 1e-04
At5g52550 28.9 2.5
> Hs11225260
Length=765
Score = 52.0 bits (123), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 22/30 (73%), Positives = 28/30 (93%), Gaps = 0/30 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQELQKL 33
HL+ LM GL+AK+FRTYNASITLQQ+L++L
Sbjct 576 HLQDLMEGLTAKVFRTYNASITLQQQLKEL 605
> Hs16418461
Length=601
Score = 50.4 bits (119), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 21/30 (70%), Positives = 28/30 (93%), Gaps = 0/30 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQELQKL 33
HL++LM GL+AK+FRTYNASITLQ++L+ L
Sbjct 412 HLQELMDGLTAKVFRTYNASITLQEQLRAL 441
> At5g55300
Length=916
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 20/27 (74%), Positives = 25/27 (92%), Gaps = 0/27 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQEL 30
HLK+L+PGL+AK+FRTYNASITL + L
Sbjct 722 HLKELVPGLTAKVFRTYNASITLDEML 748
> 7293054
Length=974
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 21/30 (70%), Positives = 25/30 (83%), Gaps = 0/30 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQELQKL 33
HLK+LM GL+AK+FRTYNAS TLQ +L L
Sbjct 800 HLKELMEGLTAKVFRTYNASKTLQSQLDLL 829
> At5g55310
Length=917
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/29 (72%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQELQK 32
HLK+LM GL+AK+FRTYNASITL L K
Sbjct 720 HLKELMAGLTAKVFRTYNASITLDLMLSK 748
> SPBC1703.14c
Length=814
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 23/41 (56%), Positives = 29/41 (70%), Gaps = 4/41 (9%)
Query 1 STN----HLKQLMPGLSAKMFRTYNASITLQQELQKLQPTL 37
STN +L LM GLSAK+FRTYNAS T+ +EL+K+ L
Sbjct 543 STNSLNKYLTSLMDGLSAKVFRTYNASYTMAEELKKMPKNL 583
> CE12282
Length=806
Score = 44.3 bits (103), Expect = 5e-05, Method: Composition-based stats.
Identities = 21/31 (67%), Positives = 27/31 (87%), Gaps = 0/31 (0%)
Query 3 NHLKQLMPGLSAKMFRTYNASITLQQELQKL 33
+HL+ LM GL+ K+FRTYNASITLQ++L KL
Sbjct 624 DHLRSLMDGLTVKVFRTYNASITLQEQLIKL 654
> CE12284
Length=734
Score = 44.3 bits (103), Expect = 6e-05, Method: Composition-based stats.
Identities = 21/31 (67%), Positives = 27/31 (87%), Gaps = 0/31 (0%)
Query 3 NHLKQLMPGLSAKMFRTYNASITLQQELQKL 33
+HL+ LM GL+ K+FRTYNASITLQ++L KL
Sbjct 552 DHLRSLMDGLTVKVFRTYNASITLQEQLIKL 582
> YOL006c
Length=769
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 17/27 (62%), Positives = 23/27 (85%), Gaps = 0/27 (0%)
Query 4 HLKQLMPGLSAKMFRTYNASITLQQEL 30
+L+ MPGL+AK+FRTYNAS T+Q +L
Sbjct 503 YLQNYMPGLTAKVFRTYNASKTMQDQL 529
> At5g52550
Length=197
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 34 QPTLRKFKEEVEREGLKKVGKAKAKKVKEEPAAAAAAAATA 74
QP +RK +++ ERE L+++ +A+ KK + E + A +AA A
Sbjct 12 QPIVRKARKKKEREELERIKQAERKKRRIEKSIATSAAIRA 52
Lambda K H
0.311 0.120 0.313
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40