bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
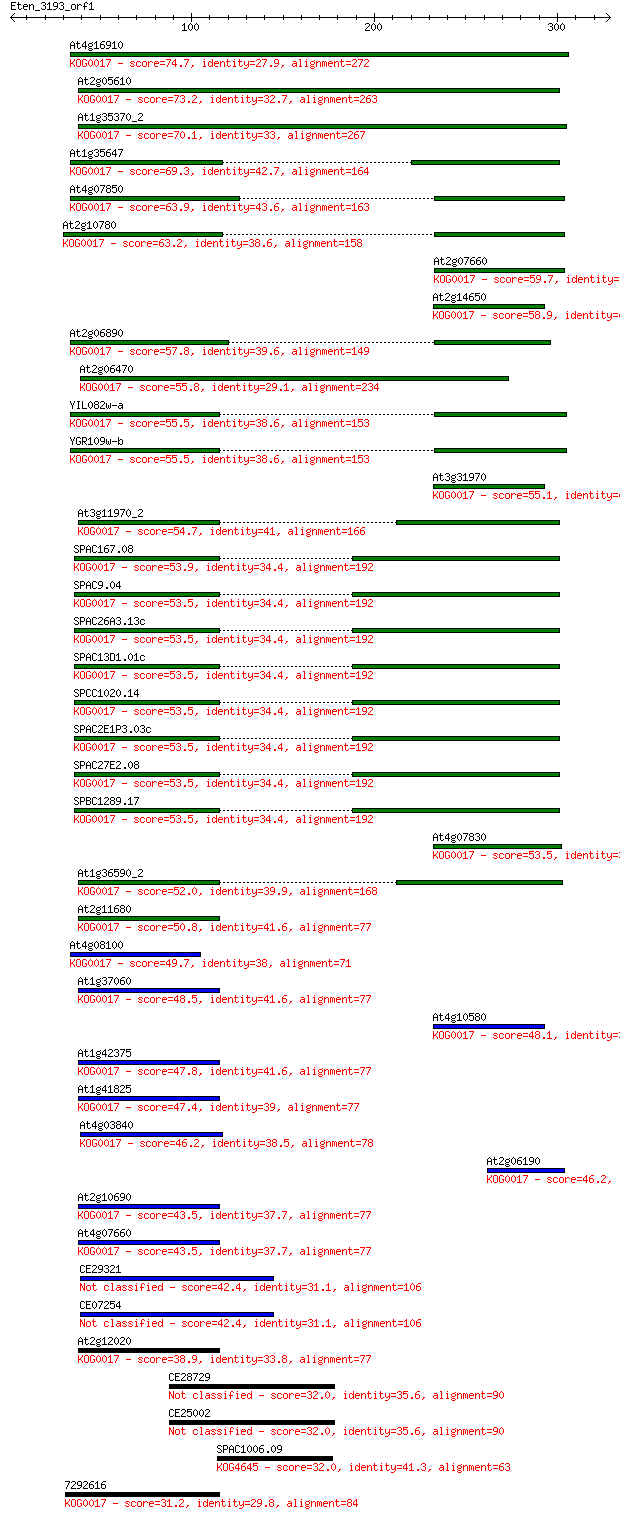
Query= Eten_3193_orf1
Length=328
Score E
Sequences producing significant alignments: (Bits) Value
At4g16910 74.7 2e-13
At2g05610 73.2 8e-13
At1g35370_2 70.1 7e-12
At1g35647 69.3 1e-11
At4g07850 63.9 4e-10
At2g10780 63.2 8e-10
At2g07660 59.7 8e-09
At2g14650 58.9 2e-08
At2g06890 57.8 3e-08
At2g06470 55.8 1e-07
YIL082w-a 55.5 2e-07
YGR109w-b 55.5 2e-07
At3g31970 55.1 2e-07
At3g11970_2 54.7 3e-07
SPAC167.08 53.9 5e-07
SPAC9.04 53.5 5e-07
SPAC26A3.13c 53.5 5e-07
SPAC13D1.01c 53.5 5e-07
SPCC1020.14 53.5 6e-07
SPAC2E1P3.03c 53.5 6e-07
SPAC27E2.08 53.5 6e-07
SPBC1289.17 53.5 6e-07
At4g07830 53.5 7e-07
At1g36590_2 52.0 2e-06
At2g11680 50.8 4e-06
At4g08100 49.7 8e-06
At1g37060 48.5 2e-05
At4g10580 48.1 2e-05
At1g42375 47.8 3e-05
At1g41825 47.4 4e-05
At4g03840 46.2 9e-05
At2g06190 46.2 1e-04
At2g10690 43.5 6e-04
At4g07660 43.5 6e-04
CE29321 42.4 0.002
CE07254 42.4 0.002
At2g12020 38.9 0.014
CE28729 32.0 1.9
CE25002 32.0 2.0
SPAC1006.09 32.0 2.1
7292616 31.2 3.2
> At4g16910
Length=687
Score = 74.7 bits (182), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 76/294 (25%), Positives = 133/294 (45%), Gaps = 35/294 (11%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKP-LRGRTARWLDILA 92
P E+ A+V AL W L + V +TDH++L ++ ++P L R RW+ ++A
Sbjct 84 PTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKYI----FTQPELNLRQRRWIKLVA 139
Query 93 KFPGFHITYVQRTRNQVADALSCRLGLPNAGIRSRRPRRRSLSSAPSPSLPEPNPEAEHP 152
+ +I Y NQV DALS RL + + R + ++ + L + E E
Sbjct 140 DYD-LNIAYHPGKANQVVDALS-RL---RTVVEAERNQVNLVNMMGTLHLNALSKEVEPL 194
Query 153 TLTTMTPAD-----------PPDVRQWPQ----AYSKCPVFRVPHKAAANQPGE-ALQIE 196
L AD +++ W Q Y + P + AL+ E
Sbjct 195 GLRAANQADLLSRIRSAQERDEEIKGWAQNNKTEYQSSNNGTIVVNGRVCGPNDKALKEE 254
Query 197 FCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLS---QVCR-ASKSLNQKPAGLLQQLLI 252
++ +H + ++R + ++ V R +K +Q P+G+LQ L I
Sbjct 255 ILKEAHQSKFS---IHPGSNKMYRDLKRYYHWVGMKKDVARWVAKEEHQVPSGMLQNLPI 311
Query 253 PSCRWAHVGLDFITDLPL-TTTGHDSIFVMVDSLSKMAHFVPTSNLTNASGIVC 305
P +W H+ +DF+T LP + H++++V+VD L+K AHF+ S+ +A+ I+
Sbjct 312 PEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISD-KDAAEIIA 364
> At2g05610
Length=780
Score = 73.2 bits (178), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 86/282 (30%), Positives = 117/282 (41%), Gaps = 38/282 (13%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA++ + KW H L S TD ++L +L + + P++ +WL L +F +
Sbjct 328 KELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQ---QQWLPKLLEFD-Y 383
Query 98 HITYVQRTRNQVADALSCRLGLPNAGIRSRRPRRRSLSSAPSPSLPE------PNPEAEH 151
I Y Q N VADALS + +LS L E + + +
Sbjct 384 EIQYKQGKENLVADALS--------RVEGSEVLHMALSVVECDLLKEIQAGYVTDGDIQG 435
Query 152 PTLTTMTPADPPDVRQWPQAYSKCPVFRVPHKAAA-NQPG---EALQIEFCNRQFTFRYV 207
AD W Q V R +K N G L+ C+
Sbjct 436 IITILQQQADSKKHYTWSQG-----VLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSGK 490
Query 208 EPYLHIRVHGL--WRICVP-FPEFLSQ--VCRASKSLNQKPAGLLQQLLIPSCRWAHVGL 262
E H RV GL W+ V F+ C+ KS N GLLQ L IP W+ V +
Sbjct 491 E-VTHQRVKGLFYWKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSM 549
Query 263 DFITDLPLTTTGHDSIFVMVDSLSKMAHFV----PTSNLTNA 300
DFI LPL + G I V+VD LSK AHF+ P S +T A
Sbjct 550 DFIDGLPL-SNGKTVIMVVVDRLSKAAHFIALAHPYSAMTVA 590
> At1g35370_2
Length=923
Score = 70.1 bits (170), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 88/283 (31%), Positives = 117/283 (41%), Gaps = 32/283 (11%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA + A+ KW H L S TD ++L +L + + P++ +WL L +F +
Sbjct 384 KELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQ---QQWLPKLLEFD-Y 439
Query 98 HITYVQRTRNQVADALSCRLGLPNAGIRSRRPRRRSLSSAPSPSLPEPNPEAE-HPTLTT 156
I Y Q N VADALS + +LS L E E L
Sbjct 440 EIQYRQGKENLVADALS--------RVEGSEVLHMALSIVECDFLKEIQVAYESDGVLKD 491
Query 157 MTPA--DPPDVRQ---WPQ-AYSKCPVFRVPHKAAANQPGEALQIEFCNRQFTFRYVEPY 210
+ A PD ++ W Q + VP+ + LQ C+ R
Sbjct 492 IISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEIT--NKLLQWLHCS-GMGGRSGRDA 548
Query 211 LHIRVHGL--WRICVP-FPEFLSQ--VCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFI 265
H RV L W+ V F+ C+ KS N GLLQ L IP W V +DFI
Sbjct 549 SHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVSMDFI 608
Query 266 TDLPLTTTGHDSIFVMVDSLSKMAHFV----PTSNLTNASGIV 304
LP + G I V+VD LSK AHFV P S LT A +
Sbjct 609 EGLP-NSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQAFL 650
> At1g35647
Length=1495
Score = 69.3 bits (168), Expect = 1e-11, Method: Composition-based stats.
Identities = 37/81 (45%), Positives = 47/81 (58%), Gaps = 6/81 (7%)
Query 220 RICVPFPEFLSQVCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIF 279
RIC P C+ +K+ +Q P GL L IPS W + +DF+ LP T TG DSIF
Sbjct 1032 RICERCP-----TCKQAKAKSQ-PHGLYTPLPIPSHPWNDISMDFVVGLPRTRTGKDSIF 1085
Query 280 VMVDSLSKMAHFVPTSNLTNA 300
V+VD SKMAHF+P +A
Sbjct 1086 VVVDRFSKMAHFIPCHKTDDA 1106
Score = 53.1 bits (126), Expect = 7e-07, Method: Composition-based stats.
Identities = 33/83 (39%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P +EL ALV AL H L +TDH++L H L+ + L R ARW++ +
Sbjct 851 PTYDKELYALVRALQTGQHYLWPKEFVIHTDHESLKH---LKGQQKLNKRHARWVEFIET 907
Query 94 FPGFHITYVQRTRNQVADALSCR 116
FP + I Y + N VADALS R
Sbjct 908 FP-YVIKYKKGKDNVVADALSRR 929
> At4g07850
Length=1138
Score = 63.9 bits (154), Expect = 4e-10, Method: Composition-based stats.
Identities = 33/71 (46%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFV 292
C+ +K+ +Q P GL L IP W + +DF+ LP T TG DSIFV+VD SKMAHF+
Sbjct 807 CKQAKAKSQ-PHGLCTPLPIPLHPWNDISMDFVVGLPRTRTGKDSIFVVVDRFSKMAHFI 865
Query 293 PTSNLTNASGI 303
P +A I
Sbjct 866 PCHKTDDAMHI 876
Score = 58.9 bits (141), Expect = 2e-08, Method: Composition-based stats.
Identities = 38/102 (37%), Positives = 52/102 (50%), Gaps = 14/102 (13%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P +EL ALV AL W H L +TDH++L H L+ + L R ARW++ +
Sbjct 662 PTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKH---LKGQQKLNKRHARWVEFIET 718
Query 94 FPGFHITYVQRTRNQVADALS----------CRLGLPNAGIR 125
FP + I Y + N VADALS RL +PN+ +R
Sbjct 719 FP-YVIKYKKGKDNVVADALSRRHEGFLFYDNRLCIPNSSLR 759
> At2g10780
Length=1611
Score = 63.2 bits (152), Expect = 8e-10, Method: Composition-based stats.
Identities = 30/72 (41%), Positives = 47/72 (65%), Gaps = 1/72 (1%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPL-TTTGHDSIFVMVDSLSKMAHF 291
C+ K+ +Q P+GLLQ L IP +W H+ +DF+T LP + H++++V+VD L+K AHF
Sbjct 1208 CQLVKAEHQVPSGLLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHF 1267
Query 292 VPTSNLTNASGI 303
+ S+ A I
Sbjct 1268 MAISDKDGAEII 1279
Score = 43.5 bits (101), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 10/88 (11%)
Query 30 PSHEPHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKP-LRGRTARWL 88
P+H+ E+ A+V L W L + V YTDH++L ++ ++P L R RW+
Sbjct 992 PTHD----LEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYI----FTQPELNLRQRRWM 1043
Query 89 DILAKFPGFHITYVQRTRNQVADALSCR 116
+++A + I Y NQVADALS R
Sbjct 1044 ELVADY-NLDIAYHPGKANQVADALSRR 1070
> At2g07660
Length=949
Score = 59.7 bits (143), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 46/72 (63%), Gaps = 1/72 (1%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPL-TTTGHDSIFVMVDSLSKMAHF 291
C+ K+ +Q P+GLLQ L I +W H+ +DF+T LP + H++++V+VD L+K AHF
Sbjct 600 CQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLTKSAHF 659
Query 292 VPTSNLTNASGI 303
+ S+ A I
Sbjct 660 MAISDKDGAEII 671
> At2g14650
Length=1328
Score = 58.9 bits (141), Expect = 2e-08, Method: Composition-based stats.
Identities = 27/61 (44%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 232 VCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHF 291
VC+ K+ +Q P G+LQ L IP +W + +DF+ LP++ T D+I+V+VD L+K AHF
Sbjct 952 VCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 1010
Query 292 V 292
+
Sbjct 1011 L 1011
> At2g06890
Length=1215
Score = 57.8 bits (138), Expect = 3e-08, Method: Composition-based stats.
Identities = 34/86 (39%), Positives = 46/86 (53%), Gaps = 4/86 (4%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P +EL ALV AL +W H L +TDH++L H L+ + L R ARW++ +
Sbjct 752 PTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKH---LKGQQKLNKRHARWVEFIET 808
Query 94 FPGFHITYVQRTRNQVADALSCRLGL 119
F + I Y + N VADALS R L
Sbjct 809 F-AYVIKYKKGKDNVVADALSQRYTL 833
Score = 41.2 bits (95), Expect = 0.003, Method: Composition-based stats.
Identities = 25/63 (39%), Positives = 32/63 (50%), Gaps = 3/63 (4%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFV 292
C+ +KS Q P GL L IP W + +DF+ LP TG DSIFV+ + F
Sbjct 941 CKQAKSKIQ-PNGLYTPLPIPKHPWNDISMDFVMGLP--RTGKDSIFVVYSPFQIVYGFN 997
Query 293 PTS 295
P S
Sbjct 998 PIS 1000
> At2g06470
Length=899
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 68/266 (25%), Positives = 109/266 (40%), Gaps = 47/266 (17%)
Query 39 ELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGFH 98
E+ A+V AL W L + V YTDH++L ++ +Q LR R RW++++A +
Sbjct 447 EMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYI-FIQPELNLRQR--RWMELVADYD-LD 502
Query 99 ITYVQRTRNQVADALSCRLGLPNAGIRSRRPRRRSLSSAPSPSLPEPNPEAEHPTLTTMT 158
I Y NQVADALS R + + + R + ++ + + + E E L
Sbjct 503 IAYHPGKANQVADALSRR----RSEVEAERSQVDLVNMMGTLHVNALSKEVESLGLGAAD 558
Query 159 PAD-----------PPDVRQWP-------QAYSKCPVFR-----VPHKAAANQP--GEAL 193
AD +++ W Q + + VP+ A + EA
Sbjct 559 QADLLSRIRLAQERDEEIKGWALNNKTEYQTSNNGTIVVNGRVCVPNDRALKEEILREAH 618
Query 194 QIEFC---NRQFTFRYVEPYLH----IRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGL 246
Q +F +R ++ Y H + W P C+ K+ +Q P+GL
Sbjct 619 QSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCP-------TCQLVKAEHQVPSGL 671
Query 247 LQQLLIPSCRWAHVGLDFITDLPLTT 272
LQ L IP +W H+ +DF L T
Sbjct 672 LQNLPIPEWKWDHITMDFAFQKALGT 697
> YIL082w-a
Length=1498
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 0/72 (0%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFV 292
C+ KS + GLLQ L I RW + +DF+T LP T+ + I V+VD SK AHF+
Sbjct 1171 CQLIKSHRPRLHGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFI 1230
Query 293 PTSNLTNASGIV 304
T +A+ ++
Sbjct 1231 ATRKTLDATQLI 1242
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P ELL ++ AL + ++L T TDH +L LQ ++P R R RWLD LA
Sbjct 957 PAGELELLGIIKALHHFRYMLHGKHFTLRTDHISLLSLQ--NKNEPAR-RVQRWLDDLAT 1013
Query 94 FPGFHITYVQRTRNQVADALS 114
+ F + Y+ +N VADA+S
Sbjct 1014 Y-DFTLEYLAGPKNVVADAIS 1033
> YGR109w-b
Length=1547
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/72 (38%), Positives = 41/72 (56%), Gaps = 0/72 (0%)
Query 233 CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFV 292
C+ KS + GLLQ L I RW + +DF+T LP T+ + I V+VD SK AHF+
Sbjct 1145 CQLIKSHRPRLHGLLQPLPIAEGRWLDISMDFVTGLPPTSNNLNMILVVVDRFSKRAHFI 1204
Query 293 PTSNLTNASGIV 304
T +A+ ++
Sbjct 1205 ATRKTLDATQLI 1216
Score = 47.0 bits (110), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 31/81 (38%), Positives = 44/81 (54%), Gaps = 4/81 (4%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P ELL ++ AL + ++L T TDH +L LQ ++P R R RWLD LA
Sbjct 931 PAGELELLGIIKALHHFRYMLHGKHFTLRTDHISLLSLQ--NKNEPAR-RVQRWLDDLAT 987
Query 94 FPGFHITYVQRTRNQVADALS 114
+ F + Y+ +N VADA+S
Sbjct 988 Y-DFTLEYLAGPKNVVADAIS 1007
> At3g31970
Length=1329
Score = 55.1 bits (131), Expect = 2e-07, Method: Composition-based stats.
Identities = 27/61 (44%), Positives = 41/61 (67%), Gaps = 1/61 (1%)
Query 232 VCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHF 291
VC+ K+ +Q P GLLQ L I +W + +DF+ LP++ T D+I+V+VD L+K AHF
Sbjct 950 VCQLVKAEHQVPGGLLQSLPILEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 1008
Query 292 V 292
+
Sbjct 1009 L 1009
> At3g11970_2
Length=958
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 50/98 (51%), Gaps = 10/98 (10%)
Query 212 HIRVHGL--WRICVPFPEFLSQVCRASKSLNQKPA---GLLQQLLIPSCRWAHVGLDFIT 266
H RV GL W+ + + + C + PA GLLQ L IP W+ V +DFI
Sbjct 562 HQRVKGLFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIE 621
Query 267 DLPLTTTGHDSIFVMVDSLSKMAHFV----PTSNLTNA 300
LP+ + G I V+VD LSK AHF+ P S LT A
Sbjct 622 GLPV-SGGKTVIMVVVDRLSKAAHFIALSHPYSALTVA 658
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA++ A+ KW H L S TD ++L +L + + P++ +WL L +F +
Sbjct 396 KELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQ---QQWLPKLLEF-DY 451
Query 98 HITYVQRTRNQVADALS 114
I Y Q N VADALS
Sbjct 452 EIQYRQGKENVVADALS 468
> SPAC167.08
Length=1214
Score = 53.9 bits (128), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 808 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 855
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 856 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 907
Score = 43.5 bits (101), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 633 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 689
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 690 QDF-NFEINYRPGSANHIADALS 711
> SPAC9.04
Length=1333
Score = 53.5 bits (127), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPAC26A3.13c
Length=1333
Score = 53.5 bits (127), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPAC13D1.01c
Length=1333
Score = 53.5 bits (127), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPCC1020.14
Length=1333
Score = 53.5 bits (127), Expect = 6e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPAC2E1P3.03c
Length=1333
Score = 53.5 bits (127), Expect = 6e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPAC27E2.08
Length=1333
Score = 53.5 bits (127), Expect = 6e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 7e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> SPBC1289.17
Length=1333
Score = 53.5 bits (127), Expect = 6e-07, Method: Composition-based stats.
Identities = 35/113 (30%), Positives = 53/113 (46%), Gaps = 13/113 (11%)
Query 188 QPGEALQIEFCNRQFTFRYVEPYLHIRVHGLWRICVPFPEFLSQVCRASKSLNQKPAGLL 247
PG L R+FT++ + + V C+ +KS N KP G L
Sbjct 927 HPGIELLTNIILRRFTWKGIRKQIQEYVQN------------CHTCQINKSRNHKPYGPL 974
Query 248 QQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNA 300
Q + W + +DFIT LP ++G++++FV+VD SKMA VP + A
Sbjct 975 QPIPPSERPWESLSMDFITALP-ESSGYNALFVVVDRFSKMAILVPCTKSITA 1026
Score = 43.1 bits (100), Expect = 8e-04, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 45/83 (54%), Gaps = 8/83 (9%)
Query 36 ATQELLALVTALDKWSHLLRVSTVTAY---TDHQALTHLQRL-QASKPLRGRTARWLDIL 91
+ +E+LA++ +L W H L ST+ + TDH+ L + R+ S+P R ARW L
Sbjct 752 SDKEMLAIIKSLKHWRHYLE-STIEPFKILTDHRNL--IGRITNESEPENKRLARWQLFL 808
Query 92 AKFPGFHITYVQRTRNQVADALS 114
F F I Y + N +ADALS
Sbjct 809 QDF-NFEINYRPGSANHIADALS 830
> At4g07830
Length=611
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 27/70 (38%), Positives = 41/70 (58%), Gaps = 1/70 (1%)
Query 232 VCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHF 291
VC+ K +Q LLQ L IP +W + +DF+ LP++ T D+I+V+VD L+K AHF
Sbjct 217 VCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRT-KDAIWVIVDRLTKSAHF 275
Query 292 VPTSNLTNAS 301
+ A+
Sbjct 276 LAIRKTDGAA 285
> At1g36590_2
Length=958
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 38/100 (38%), Positives = 49/100 (49%), Gaps = 10/100 (10%)
Query 212 HIRVHGLWRICVPFPEFLSQV-----CRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFIT 266
H RV GL+ + + + C+ KS GLLQ L IP W+ V +DFI
Sbjct 562 HQRVKGLFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIE 621
Query 267 DLPLTTTGHDSIFVMVDSLSKMAHFV----PTSNLTNASG 302
LP+ + G I V+VD LSK AHF+ P S LT A
Sbjct 622 GLPV-SGGKTVIMVVVDRLSKAAHFIALSHPYSALTVAQA 660
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA++ A+ KW H L S TD ++L +L + + P++ +WL L +F +
Sbjct 396 KELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQ---QQWLPKLLEF-DY 451
Query 98 HITYVQRTRNQVADALS 114
I Y Q N VADALS
Sbjct 452 EIQYRQGKENVVADALS 468
> At2g11680
Length=301
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 32/77 (41%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA+V A +K+ L S VT YTDH AL H+ + +KP R RW+ +L +F
Sbjct 150 KELLAVVFAFEKFRSYLVGSKVTVYTDHAALRHIYAKKDTKP---RLLRWILLLQEFD-M 205
Query 98 HITYVQRTRNQVADALS 114
I + N VAD LS
Sbjct 206 EIVDKKGIENGVADHLS 222
> At4g08100
Length=1054
Score = 49.7 bits (117), Expect = 8e-06, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 4/71 (5%)
Query 34 PHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAK 93
P +EL ALV AL W H L +TDH++L H L+ + L R ARW++ +
Sbjct 775 PTYDKELYALVRALQTWQHYLWPKKFVIHTDHESLKH---LKGQQKLNKRHARWVEFIET 831
Query 94 FPGFHITYVQR 104
FP + I Y +R
Sbjct 832 FP-YVIKYKKR 841
> At1g37060
Length=1734
Score = 48.5 bits (114), Expect = 2e-05, Method: Composition-based stats.
Identities = 32/77 (41%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA+V A +K+ L S VT YTDH AL H+ + +KP R RW+ +L +F
Sbjct 1231 KELLAVVFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKP---RLLRWILLLQEF-DM 1286
Query 98 HITYVQRTRNQVADALS 114
I + N VAD LS
Sbjct 1287 EIVDKKGIENGVADHLS 1303
> At4g10580
Length=1240
Score = 48.1 bits (113), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/61 (39%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Query 232 VCRASKSLNQKPAGLLQQLLIPSCRWAHVGLDFITDLPLTTTGHDSIFVMVDSLSKMAHF 291
VC+ K+ +Q G+LQ L IP +W + +D + L ++ T D+I+V+VD L+K AHF
Sbjct 987 VCQLVKAEHQVLGGMLQSLPIPEWKWDFITMDLVVGLRVSRT-KDAIWVIVDRLTKSAHF 1045
Query 292 V 292
+
Sbjct 1046 L 1046
> At1g42375
Length=1773
Score = 47.8 bits (112), Expect = 3e-05, Method: Composition-based stats.
Identities = 32/77 (41%), Positives = 42/77 (54%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA+V A +K+ L S VT YTDH AL HL + +KP R RW+ +L +F
Sbjct 1228 KELLAVVFAFEKFRSYLVGSKVTVYTDHAALRHLYAKKDTKP---RLLRWILLLQEF-DM 1283
Query 98 HITYVQRTRNQVADALS 114
I + N AD LS
Sbjct 1284 EIVDKKGIENGAADHLS 1300
> At1g41825
Length=884
Score = 47.4 bits (111), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 43/77 (55%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA+V A DK+ L S V +TDH AL +L + + +KP R RW+ +L +F
Sbjct 787 KELLAVVFAFDKFRSYLVGSKVIVHTDHSALKYLMQKKDAKP---RLLRWILLLQEF-DI 842
Query 98 HITYVQRTRNQVADALS 114
+ + N VAD LS
Sbjct 843 EVRDRKGIENGVADHLS 859
> At4g03840
Length=973
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 30/79 (37%), Positives = 44/79 (55%), Gaps = 6/79 (7%)
Query 39 ELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKP-LRGRTARWLDILAKFPGF 97
E+ A+V AL W L + V YTDH++L ++ ++P L R RW++++A +
Sbjct 447 EMAAVVFALKIWRSYLYGAKVQIYTDHKSLKYI----FTQPELNLRQRRWMELVADYD-L 501
Query 98 HITYVQRTRNQVADALSCR 116
I Y NQVADALS R
Sbjct 502 DIAYHAGKANQVADALSRR 520
> At2g06190
Length=280
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 262 LDFITDLPLTTTGHDSIFVMVDSLSKMAHFVPTSNLTNASGI 303
+DF+ LP T G DS+FV+VD SKM HF+ +AS I
Sbjct 1 MDFVLGLPRTQRGVDSVFVVVDRFSKMTHFIACKKTADASNI 42
> At2g10690
Length=622
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 38/77 (49%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELLA+V A +K L V Y DH AL H + A K + R RW+ +L +F
Sbjct 395 KELLAVVFAFEKIRSYLVGFKVKVYIDHAALRH---IYAKKETKSRLLRWILLLQEFD-M 450
Query 98 HITYVQRTRNQVADALS 114
I + N VAD LS
Sbjct 451 EIIDKKGVENGVADHLS 467
> At4g07660
Length=724
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 29/77 (37%), Positives = 39/77 (50%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+ELL +V A K+ L S VT YTDH AL H+ + +KP R R + +L +F
Sbjct 617 KELLVVVFAFKKFRSYLVESKVTVYTDHAALRHMYAKKDTKP---RLLRGILLLQEFD-M 672
Query 98 HITYVQRTRNQVADALS 114
I + N AD LS
Sbjct 673 EIVEKKGIENGAADHLS 689
> CE29321
Length=2186
Score = 42.4 bits (98), Expect = 0.002, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 53/107 (49%), Gaps = 6/107 (5%)
Query 39 ELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGFH 98
E LA++ AL ++ ++ + +T +TDH+ L L + PL R RW + +F
Sbjct 1291 EALAMMFALRRFKTIIYGTAITVFTDHKPLISLLK---GSPLADRLWRWSIEILEF-DVK 1346
Query 99 ITYVQRTRNQVADALSCRLGLPNAGIRSRRPRR-RSLSSAPSPSLPE 144
I Y+ N VADALS R G P + + + S+ +A LP+
Sbjct 1347 IVYLAGKANAVADALS-RGGCPPNELEEEQTKELTSIVNAIQTELPD 1392
> CE07254
Length=2175
Score = 42.4 bits (98), Expect = 0.002, Method: Composition-based stats.
Identities = 33/107 (30%), Positives = 53/107 (49%), Gaps = 6/107 (5%)
Query 39 ELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGFH 98
E LA++ AL ++ ++ + +T +TDH+ L L + PL R RW + +F
Sbjct 1280 EALAMMFALRRFKTIIYGTAITVFTDHKPLISLLK---GSPLADRLWRWSIEILEF-DVK 1335
Query 99 ITYVQRTRNQVADALSCRLGLPNAGIRSRRPRR-RSLSSAPSPSLPE 144
I Y+ N VADALS R G P + + + S+ +A LP+
Sbjct 1336 IVYLAGKANAVADALS-RGGCPPNELEEEQTKELTSIVNAIQTELPD 1381
> At2g12020
Length=976
Score = 38.9 bits (89), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 40/77 (51%), Gaps = 4/77 (5%)
Query 38 QELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDILAKFPGF 97
+E LA+V A +K+ L V +TDH AL +L + + +KP R RW+ +L +F
Sbjct 286 KEFLAVVFAFEKFRSYLVGLKVIVHTDHAALKYLMQNKDAKP---RLLRWILLLQEF-DI 341
Query 98 HITYVQRTRNQVADALS 114
+ + VAD LS
Sbjct 342 EVRDKKGVEKCVADHLS 358
> CE28729
Length=4203
Score = 32.0 bits (71), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 50/102 (49%), Gaps = 13/102 (12%)
Query 88 LDILAKFPGFHITYVQRTRNQVADAL-SCRLG-LPNAGIRS---------RRPRRRSLSS 136
L+I +P + Y+ R N+V +A+ S +L P GI S RR +
Sbjct 2786 LEISPTYPEDNGEYICRAVNRVGEAVTSTKLTCTPKEGIISATQLPERMANAGRRIAEIE 2845
Query 137 APSPSLPEPNPEAEH-PTLTTMTPADPPDVRQWPQAYSKCPV 177
AP P+ E P+A+H P T A PP++++ QA+ +C V
Sbjct 2846 APRPAR-EDAPDADHGPPKFTSALAGPPELQEGQQAHLECQV 2886
> CE25002
Length=4927
Score = 32.0 bits (71), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 50/102 (49%), Gaps = 13/102 (12%)
Query 88 LDILAKFPGFHITYVQRTRNQVADAL-SCRLG-LPNAGIRS---------RRPRRRSLSS 136
L+I +P + Y+ R N+V +A+ S +L P GI S RR +
Sbjct 3546 LEISPTYPEDNGEYICRAVNRVGEAVTSTKLTCTPKEGIISATQLPERMANAGRRIAEIE 3605
Query 137 APSPSLPEPNPEAEH-PTLTTMTPADPPDVRQWPQAYSKCPV 177
AP P+ E P+A+H P T A PP++++ QA+ +C V
Sbjct 3606 APRPAR-EDAPDADHGPPKFTSALAGPPELQEGQQAHLECQV 3646
> SPAC1006.09
Length=699
Score = 32.0 bits (71), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 32/74 (43%), Gaps = 11/74 (14%)
Query 114 SCRLGLPNAGIRS--RRPR------RRSLSSAPSPSLPEPNPEAEHP---TLTTMTPADP 162
S L LPN G RRP RS +SA E AE P LT TP DP
Sbjct 53 SSMLELPNNGKEENHRRPSVARSSSDRSKASAKEDLFSEAFRMAEQPPAEALTISTPVDP 112
Query 163 PDVRQWPQAYSKCP 176
++ + +AY+ P
Sbjct 113 INIDELDRAYAVSP 126
> 7292616
Length=1062
Score = 31.2 bits (69), Expect = 3.2, Method: Composition-based stats.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 4/84 (4%)
Query 31 SHEPHATQELLALVTALDKWSHLLRVSTVTAYTDHQALTHLQRLQASKPLRGRTARWLDI 90
+H +ELLA+V A+ + L + TDH+ L L+ L+ + RW
Sbjct 525 THYATIKKELLAIVWAVKYFRPYLFGNKFLLVTDHKPLVWLKNLKEPNSM---LVRWKLQ 581
Query 91 LAKFPGFHITYVQRTRNQVADALS 114
L ++ + Y + ++N VADALS
Sbjct 582 LLEY-DCEVIYKKGSQNVVADALS 604
Lambda K H
0.322 0.134 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7707093114
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40