bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3220_orf3
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
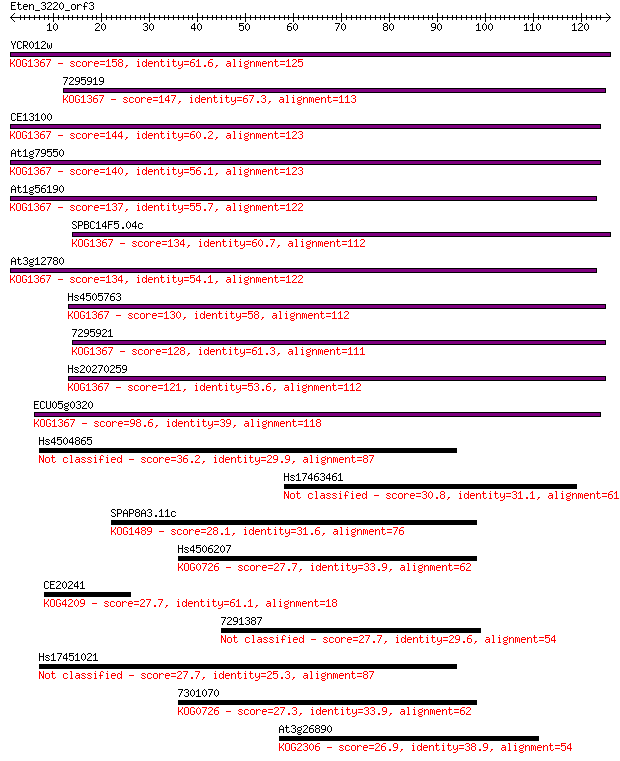
YCR012w 158 2e-39
7295919 147 4e-36
CE13100 144 5e-35
At1g79550 140 8e-34
At1g56190 137 6e-33
SPBC14F5.04c 134 3e-32
At3g12780 134 4e-32
Hs4505763 130 6e-31
7295921 128 3e-30
Hs20270259 121 3e-28
ECU05g0320 98.6 2e-21
Hs4504865 36.2 0.015
Hs17463461 30.8 0.59
SPAP8A3.11c 28.1 3.6
Hs4506207 27.7 5.1
CE20241 27.7 5.3
7291387 27.7 5.5
Hs17451021 27.7 5.6
7301070 27.3 6.9
At3g26890 26.9 8.2
> YCR012w
Length=416
Score = 158 bits (400), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 77/125 (61%), Positives = 96/125 (76%), Gaps = 0/125 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
+ADANT+ DK G+P GW GLD GP++ +L A +++AKTIVWNGPPGVFE FA G+
Sbjct 291 SADANTKTVTDKEGIPAGWQGLDNGPESRKLFAATVAKAKTIVWNGPPGVFEFEKFAAGT 350
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A + V ++AAG T I+GGGDTA++ +K G+ K+SHVSTGGGASLELLEGKELPGVA
Sbjct 351 KALLDEVVKSSAAGNTVIIGGGDTATVAKKYGVTDKISHVSTGGGASLELLEGKELPGVA 410
Query 121 ALSSK 125
LS K
Sbjct 411 FLSEK 415
> 7295919
Length=415
Score = 147 bits (371), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 76/113 (67%), Positives = 85/113 (75%), Gaps = 0/113 (0%)
Query 12 KAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAAT 71
+AG+PDG MGLDVGPKT EL A I+RAK IVWNGPPGVFE P FA G+ + + V AAT
Sbjct 302 EAGIPDGHMGLDVGPKTRELFAAPIARAKLIVWNGPPGVFEFPNFANGTKSIMDGVVAAT 361
Query 72 AAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G SI+GGGDTAS K +SHVSTGGGASLELLEGK LPGVAAL+S
Sbjct 362 KNGTVSIIGGGDTASCCAKWNTEALVSHVSTGGGASLELLEGKTLPGVAALTS 414
> CE13100
Length=417
Score = 144 bits (362), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 74/123 (60%), Positives = 89/123 (72%), Gaps = 0/123 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DA ++ + GVPDG MGLDVGP++ ++ A I RAKTIVWNGP GVFE FA G+
Sbjct 292 AEDATSKTVTAEEGVPDGHMGLDVGPESSKIFAAAIQRAKTIVWNGPAGVFEFDKFATGT 351
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
+ + V ATAAGA +I+GGGDTA+ +K K+SHVSTGGGASLELLEGK LPGV
Sbjct 352 KSLMDEVVKATAAGAITIIGGGDTATAAKKYNTEDKVSHVSTGGGASLELLEGKVLPGVD 411
Query 121 ALS 123
ALS
Sbjct 412 ALS 414
> At1g79550
Length=401
Score = 140 bits (352), Expect = 8e-34, Method: Compositional matrix adjust.
Identities = 69/123 (56%), Positives = 88/123 (71%), Gaps = 1/123 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I A +PDGWMGLD+GP +++ + KTI+WNGP GVFE FA G+
Sbjct 278 APDANSKIVPATA-IPDGWMGLDIGPDSIKTFSEALDTTKTIIWNGPMGVFEFDKFAAGT 336
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A+ +A + G T+I+GGGD+ + VEK G+A KMSH+STGGGASLELLEGK LPGV
Sbjct 337 EAVAKQLAELSGKGVTTIIGGGDSVAAVEKVGLADKMSHISTGGGASLELLEGKPLPGVL 396
Query 121 ALS 123
AL
Sbjct 397 ALD 399
> At1g56190
Length=478
Score = 137 bits (344), Expect = 6e-33, Method: Compositional matrix adjust.
Identities = 68/122 (55%), Positives = 87/122 (71%), Gaps = 1/122 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I A +PDGWMGLD+GP +V+ + +T++WNGP GVFE FAKG+
Sbjct 350 APDANSKIVPASA-IPDGWMGLDIGPDSVKTFNEALDTTQTVIWNGPMGVFEFEKFAKGT 408
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A +A + G T+I+GGGD+ + VEK G+A MSH+STGGGASLELLEGK LPGV
Sbjct 409 EAVANKLAELSKKGVTTIIGGGDSVAAVEKVGVAGVMSHISTGGGASLELLEGKVLPGVV 468
Query 121 AL 122
AL
Sbjct 469 AL 470
> SPBC14F5.04c
Length=414
Score = 134 bits (338), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 68/112 (60%), Positives = 80/112 (71%), Gaps = 0/112 (0%)
Query 14 GVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAA 73
G+PDGWMGLD GPK+ +I+ +KTIVWNGP GVFE FAKG+ + +A A
Sbjct 303 GIPDGWMGLDCGPKSSAKFAEVITTSKTIVWNGPAGVFEFDNFAKGTKSMLDACVKTCEA 362
Query 74 GATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSSK 125
G IVGGGDTA++ +K G +SHVSTGGGASLELLEGK LPGV ALSSK
Sbjct 363 GNVVIVGGGDTATVAKKYGKEDALSHVSTGGGASLELLEGKALPGVVALSSK 414
> At3g12780
Length=481
Score = 134 bits (337), Expect = 4e-32, Method: Compositional matrix adjust.
Identities = 66/122 (54%), Positives = 87/122 (71%), Gaps = 1/122 (0%)
Query 1 AADANTQICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGS 60
A DAN++I +G+ DGWMGLD+GP +++ + +T++WNGP GVFEM FA G+
Sbjct 353 APDANSKIVP-ASGIEDGWMGLDIGPDSIKTFNEALDTTQTVIWNGPMGVFEMEKFAAGT 411
Query 61 TAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVA 120
A A +A + G T+I+GGGD+ + VEK G+A MSH+STGGGASLELLEGK LPGV
Sbjct 412 EAIANKLAELSEKGVTTIIGGGDSVAAVEKVGVAGVMSHISTGGGASLELLEGKVLPGVI 471
Query 121 AL 122
AL
Sbjct 472 AL 473
> Hs4505763
Length=417
Score = 130 bits (327), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 65/112 (58%), Positives = 81/112 (72%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
+G+P GWMGLD GP++ + ++RAK IVWNGP GVFE AFA+G+ A + V AT+
Sbjct 305 SGIPAGWMGLDCGPESSKKYAEAVTRAKQIVWNGPVGVFEWEAFARGTKALMDEVVKATS 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +I+GGGDTA+ K K+SHVSTGGGASLELLEGK LPGV ALS+
Sbjct 365 RGCITIIGGGDTATCCAKWNTEDKVSHVSTGGGASLELLEGKVLPGVDALSN 416
> 7295921
Length=389
Score = 128 bits (321), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 68/111 (61%), Positives = 81/111 (72%), Gaps = 0/111 (0%)
Query 14 GVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAA 73
G+P+ MGLD G + ++ I +KTIVWNGPPG+FE F+KG+ + EAV AT
Sbjct 278 GIPEDMMGLDHGQASSQMFAGAICASKTIVWNGPPGLFENELFSKGTESMLEAVIGATNR 337
Query 74 GATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
GAT+IVGGGDTA+ K G A K+SHVSTGGGASLELLEGK LPGVAALS
Sbjct 338 GATTIVGGGDTATACLKFGGADKVSHVSTGGGASLELLEGKVLPGVAALSD 388
> Hs20270259
Length=417
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 60/112 (53%), Positives = 80/112 (71%), Gaps = 0/112 (0%)
Query 13 AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATA 72
+G+ GWMGLD GP++ + ++++A+ IVWNGP GVFE AFAKG+ A + + AT+
Sbjct 305 SGISPGWMGLDCGPESNKNHAQVVAQARLIVWNGPLGVFEWDAFAKGTKALMDEIVKATS 364
Query 73 AGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAALSS 124
G +++GGGDTA+ K K+SHVSTG GASLELLEGK LPGV ALS+
Sbjct 365 KGCITVIGGGDTATCCAKWNTEDKVSHVSTGRGASLELLEGKILPGVEALSN 416
> ECU05g0320
Length=388
Score = 98.6 bits (244), Expect = 2e-21, Method: Composition-based stats.
Identities = 46/122 (37%), Positives = 73/122 (59%), Gaps = 4/122 (3%)
Query 6 TQICEDK----AGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGST 61
T +C D + +P +D+GP+T+E+ + +S+AK + WNGPPG++E +F++G+
Sbjct 264 TVVCNDSVQTTSNIPPEGSAMDIGPETIEMLRREVSKAKAVFWNGPPGMYEEQSFSRGTE 323
Query 62 AFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMSHVSTGGGASLELLEGKELPGVAA 121
A A+ + G S GGG+TA + G ++VSTGGG ++LL GK LPG+
Sbjct 324 ALVSALESLKGEGKKSFCGGGETAGAIRLFGSYDAFTYVSTGGGVLMKLLGGKRLPGLDF 383
Query 122 LS 123
LS
Sbjct 384 LS 385
> Hs4504865
Length=711
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 26/87 (29%), Positives = 38/87 (43%), Gaps = 0/87 (0%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
QI D G+P+ + L P++V+ AK M+ + GPPG F A + E
Sbjct 179 QISPDSGGLPERSVSLTGAPESVQKAKMMLDDIVSRGRGGPPGQFHDNANGGQNGTVQEI 238
Query 67 VAAATAAGATSIVGGGDTASLVEKCGM 93
+ A AG GG L E+ G+
Sbjct 239 MIPAGKAGLVIGKGGETIKQLQERAGV 265
> Hs17463461
Length=279
Score = 30.8 bits (68), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 29/66 (43%), Gaps = 5/66 (7%)
Query 58 KGSTAFAEAVAAATAAGATSIVGGGDTASLVEKCGMA-----PKMSHVSTGGGASLELLE 112
+G E V S+V D + +++C P+M V GG A+L+L
Sbjct 116 EGRLVIEEVVFLEYHTACKSLVDKRDKETDLKRCDKCYDKGKPRMLWVPRGGTAALDLNN 175
Query 113 GKELPG 118
K+LPG
Sbjct 176 MKDLPG 181
> SPAP8A3.11c
Length=419
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 11/83 (13%)
Query 22 LDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAG------- 74
L V P TV + + +++ W PG + P KG +F V+ AT G
Sbjct 115 LYVPPGTVIREISAVRSEQSLEWVQMPGKTKPPKLKKGQISF---VSEATRHGKELLYYR 171
Query 75 ATSIVGGGDTASLVEKCGMAPKM 97
A+S++ G SL E+C P++
Sbjct 172 ASSMISGAAEYSL-EECDTTPQI 193
> Hs4506207
Length=440
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 13/63 (20%)
Query 36 ISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVGGGDTASLVEK-CGMA 94
I K ++ GPPG G T A+AVA T+A +VG + L++K G
Sbjct 216 IKPPKGVILYGPPGT--------GKTLLAKAVANQTSATFLRVVG----SELIQKYLGDG 263
Query 95 PKM 97
PK+
Sbjct 264 PKL 266
> CE20241
Length=437
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 11/19 (57%), Positives = 14/19 (73%), Gaps = 1/19 (5%)
Query 8 ICEDKAGVPDGW-MGLDVG 25
+ EDK G+P GW +GLD G
Sbjct 172 VSEDKKGLPQGWLLGLDKG 190
> 7291387
Length=785
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 16/54 (29%), Positives = 23/54 (42%), Gaps = 0/54 (0%)
Query 45 NGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVGGGDTASLVEKCGMAPKMS 98
+G F +PA K S A A+ A ++ G + S +E C M P S
Sbjct 193 SGLLAAFSLPAIQKTSAAGADGPAHEVVELTPALHSIGISRSFIEHCSMTPSPS 246
> Hs17451021
Length=571
Score = 27.7 bits (60), Expect = 5.6, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 37/87 (42%), Gaps = 1/87 (1%)
Query 7 QICEDKAGVPDGWMGLDVGPKTVELAKAMISRAKTIVWNGPPGVFEMPAFAKGSTAFAEA 66
++ + +G+P + L P++++ AK M+ + GPPG F A G +
Sbjct 90 RVPDGSSGLPKHSVSLTRAPESLQKAKMMLDDIVSRGHGGPPGQFHDTA-KGGQNGTVQD 148
Query 67 VAAATAAGATSIVGGGDTASLVEKCGM 93
+ A AG GG L E G+
Sbjct 149 MIPAGNAGLVIAKGGETMKQLQEYAGV 175
> 7301070
Length=439
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 30/63 (47%), Gaps = 13/63 (20%)
Query 36 ISRAKTIVWNGPPGVFEMPAFAKGSTAFAEAVAAATAAGATSIVGGGDTASLVEK-CGMA 94
I K ++ GPPG G T A+AVA T+A +VG + L++K G
Sbjct 215 IKPPKGVILYGPPGT--------GKTLLAKAVANQTSATFLRVVG----SELIQKYLGDG 262
Query 95 PKM 97
PK+
Sbjct 263 PKL 265
> At3g26890
Length=716
Score = 26.9 bits (58), Expect = 8.2, Method: Composition-based stats.
Identities = 21/57 (36%), Positives = 32/57 (56%), Gaps = 3/57 (5%)
Query 57 AKGSTAFAEAVAAATAAGATSIVG--GGDTASLVEK-CGMAPKMSHVSTGGGASLEL 110
A GST++A +++ GA IVG G+T+SL K +A S ST G A +++
Sbjct 150 AHGSTSYAAGKVSSSLTGARRIVGFASGETSSLDNKQTSVAVDHSLSSTVGVAGVDI 206
Lambda K H
0.311 0.128 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1183965632
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40