bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3224_orf1
Length=149
Score E
Sequences producing significant alignments: (Bits) Value
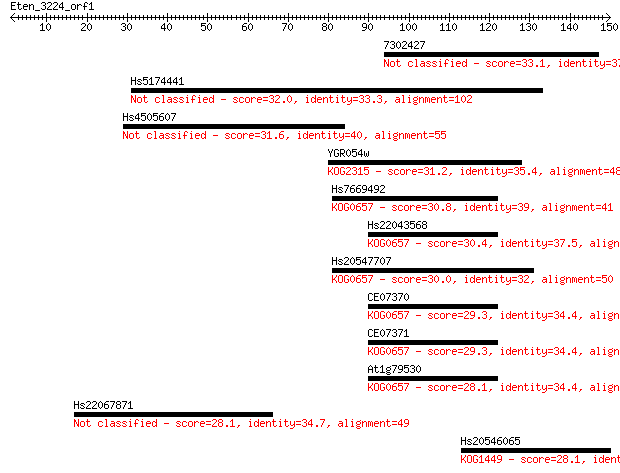
7302427 33.1 0.21
Hs5174441 32.0 0.42
Hs4505607 31.6 0.66
YGR054w 31.2 0.75
Hs7669492 30.8 0.94
Hs22043568 30.4 1.5
Hs20547707 30.0 1.6
CE07370 29.3 3.0
CE07371 29.3 3.0
At1g79530 28.1 6.8
Hs22067871 28.1 6.9
Hs20546065 28.1 7.4
> 7302427
Length=613
Score = 33.1 bits (74), Expect = 0.21, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 24/62 (38%), Gaps = 9/62 (14%)
Query 94 GGVQRAAAAVRGGSSRVVPGAEAADSPIP---------AAAPAGGVHCGAPHLVCCLLRG 144
GG Q+ +AAV SSR + D P + A C PHL CC G
Sbjct 251 GGTQKPSAAVSTASSRHNHILKKVDQPFSQKQTPRKGSSTTSASKTRCNCPHLRCCRANG 310
Query 145 AA 146
A
Sbjct 311 EA 312
> Hs5174441
Length=1530
Score = 32.0 bits (71), Expect = 0.42, Method: Composition-based stats.
Identities = 34/107 (31%), Positives = 42/107 (39%), Gaps = 23/107 (21%)
Query 31 ERRDPAADAAVDGDDGGAVQGSRHPRKLAAVLLEAAGVR-----VDGGLCCSSRRSSVRR 85
E+ PAA A DG G+R +VL++AAG GG +RR + +
Sbjct 1427 EQLPPAAQAG-DGQPAAPDGGARPDGARVSVLVDAAGASHCQPCAPGGSRRPTRRPTETQ 1485
Query 86 GEGGAGVLGGVQRAAAAVRGGSSRVVPGAEAADSPIPAAAPAGGVHC 132
E G QR A V A DSP AAAP G C
Sbjct 1486 SEQG------FQRRAGRV-----------TAVDSPPCAAAPEGSYQC 1515
> Hs4505607
Length=1627
Score = 31.6 bits (70), Expect = 0.66, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 28/65 (43%), Gaps = 13/65 (20%)
Query 29 GDERRDPAADAAVDGDDGGAVQGSRHPRKLAAVLLEAAGV----------RVDGGLCCSS 78
GDE DP + + G DGG + RHP A V + GV DGG CC
Sbjct 420 GDENCDPECNHTLTGHDGGDCRHLRHP---AFVKKQHNGVCDMDCNYERFNFDGGECCDP 476
Query 79 RRSSV 83
++V
Sbjct 477 EITNV 481
> YGR054w
Length=642
Score = 31.2 bits (69), Expect = 0.75, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 20/50 (40%), Gaps = 2/50 (4%)
Query 80 RSSVRRGEGGAGVLGGVQRAAAAVRGGSSRVVPGAE--AADSPIPAAAPA 127
R + G G GG + A R G R+VPG AA IP P
Sbjct 491 REASSNGNGSKAKAGGAYKPPHARRTGGGRIVPGVPPGAAKKTIPGLVPG 540
> Hs7669492
Length=335
Score = 30.8 bits (68), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 16/48 (33%), Positives = 29/48 (60%), Gaps = 7/48 (14%)
Query 81 SSVRRGEGGA-------GVLGGVQRAAAAVRGGSSRVVPGAEAADSPI 121
S ++ G+ GA GV +++A A ++GG+ RV+ A +AD+P+
Sbjct 83 SKIKWGDAGAEYVVESTGVFTTMEKAGAHLQGGAKRVIISAPSADAPM 130
> Hs22043568
Length=247
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 12/32 (37%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 90 AGVLGGVQRAAAAVRGGSSRVVPGAEAADSPI 121
G+ V++A A + GG+ RV+ A +AD+P+
Sbjct 47 TGIFTTVEKAGAHLEGGAKRVITSAPSADAPM 78
> Hs20547707
Length=303
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 16/57 (28%), Positives = 28/57 (49%), Gaps = 7/57 (12%)
Query 81 SSVRRGEGGA-------GVLGGVQRAAAAVRGGSSRVVPGAEAADSPIPAAAPAGGV 130
S ++ G+ G G+ ++ A A ++ G+ RV+ A +AD P+ AP V
Sbjct 83 SKIKWGDAGTEYVMESTGIFTTMENAGAHLQWGAKRVIISAPSADPPMFTMAPLAKV 139
> CE07370
Length=341
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 90 AGVLGGVQRAAAAVRGGSSRVVPGAEAADSPI 121
GV +++A A ++GG+ +V+ A +AD+P+
Sbjct 104 TGVFTTIEKANAHLKGGAKKVIISAPSADAPM 135
> CE07371
Length=341
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 90 AGVLGGVQRAAAAVRGGSSRVVPGAEAADSPI 121
GV +++A A ++GG+ +V+ A +AD+P+
Sbjct 104 TGVFTTIEKANAHLKGGAKKVIISAPSADAPM 135
> At1g79530
Length=422
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 90 AGVLGGVQRAAAAVRGGSSRVVPGAEAADSPI 121
+GV + +AA+ ++GG+ +V+ A +AD+P+
Sbjct 183 SGVFTTLSKAASHLKGGAKKVIISAPSADAPM 214
> Hs22067871
Length=630
Score = 28.1 bits (61), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 1/50 (2%)
Query 17 ISTAGSELRDAGGDERRDPAADAAVDG-DDGGAVQGSRHPRKLAAVLLEA 65
+S A S + GG + +PAADA+ G D+ G+ Q + P + A +++E
Sbjct 353 VSWAVSAVPGLGGQQSSEPAADASPPGQDEPGSYQENVPPAQKAPLIMET 402
> Hs20546065
Length=875
Score = 28.1 bits (61), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/37 (43%), Positives = 21/37 (56%), Gaps = 3/37 (8%)
Query 113 GAEAADSPIPAAAPAGGVHCGAPHLVCCLLRGAAGAL 149
GA A+ +P PA +P + PHL+ LLRGA L
Sbjct 403 GAPASATPTPALSPGRSLR---PHLIPLLLRGAEAPL 436
Lambda K H
0.320 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40