bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3229_orf1
Length=141
Score E
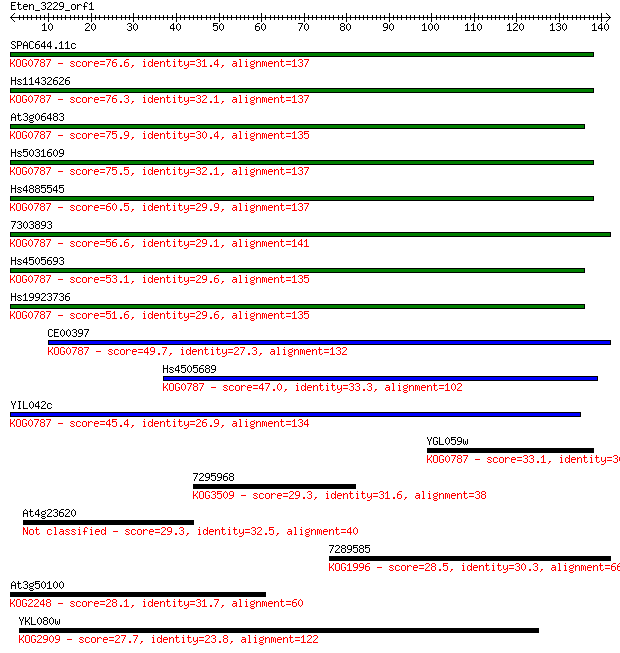
Sequences producing significant alignments: (Bits) Value
SPAC644.11c 76.6 1e-14
Hs11432626 76.3 2e-14
At3g06483 75.9 2e-14
Hs5031609 75.5 3e-14
Hs4885545 60.5 1e-09
7303893 56.6 1e-08
Hs4505693 53.1 1e-07
Hs19923736 51.6 5e-07
CE00397 49.7 2e-06
Hs4505689 47.0 1e-05
YIL042c 45.4 4e-05
YGL059w 33.1 0.19
7295968 29.3 2.3
At4g23620 29.3 2.7
7289585 28.5 4.1
At3g50100 28.1 6.2
YKL080w 27.7 7.7
> SPAC644.11c
Length=425
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 79/143 (55%), Gaps = 6/143 (4%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLS--LDVSFDFFLERFFYFRIGRRVMVNNLLA 58
F LL R +HD+ + +Q+ +R + +D S FL+RF+ RIG R+++ +A
Sbjct 167 FAYLLNTIRTRHDNVAVEIALDIQEYRRKTNQIDNSIQIFLDRFYMSRIGIRMLLGQYIA 226
Query 59 M--QNPKPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGI--APLVVVAGNLETEFATI 114
+ + P+ + G+++ R ++I A+ + C+ +YG+ AP + + + E +
Sbjct 227 LVSEPPRENYVGVISTRANIYQIIEGAAENAKYICRLAYGLFEAPEIQIICDPSLEMMYV 286
Query 115 PEHLALIITEVLKNALRATVEFY 137
HL + E+LKN+LRATVEF+
Sbjct 287 ESHLNHAVFEILKNSLRATVEFH 309
> Hs11432626
Length=412
Score = 76.3 bits (186), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 72/138 (52%), Gaps = 1/138 (0%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLDVSF-DFFLERFFYFRIGRRVMVNNLLAM 59
+ +L+ Q H V LL L++ ++ D +FL++ R+G R++ + LA+
Sbjct 151 YCQLVRQLLDDHKDVVTLLAEGLRESRKHIEDEKLVRYFLDKTLTSRLGIRMLATHHLAL 210
Query 60 QNPKPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVVAGNLETEFATIPEHLA 119
KP + GI+ R P ++I R C+ YG AP V + G++ F IP L
Sbjct 211 HEDKPDFVGIICTRLSPKKIIEKWVDFARRLCEHKYGNAPRVRINGHVAARFPFIPMPLD 270
Query 120 LIITEVLKNALRATVEFY 137
I+ E+LKNA+RAT+E +
Sbjct 271 YILPELLKNAMRATMESH 288
> At3g06483
Length=380
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 41/140 (29%), Positives = 76/140 (54%), Gaps = 5/140 (3%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLDVSFDF---FLERFFYFRIGRRVMVNNLL 57
F ++++ + +H++ V ++ + QLK+ + D FL+RF+ RIG R+++ +
Sbjct 110 FTQMIKAVKVRHNNVVPMMALGVNQLKKGMNSGNLDEIHQFLDRFYLSRIGIRMLIGQHV 169
Query 58 AMQNPKPGW--CGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVVAGNLETEFATIP 115
+ NP P G ++ + P EV +++ R C YG AP + + G+ F +P
Sbjct 170 ELHNPNPPLHTVGYIHTKMSPMEVARNASEDARSICFREYGSAPEINIYGDPSFTFPYVP 229
Query 116 EHLALIITEVLKNALRATVE 135
HL L++ E++KN+LRA E
Sbjct 230 THLHLMMYELVKNSLRAVQE 249
> Hs5031609
Length=412
Score = 75.5 bits (184), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 44/138 (31%), Positives = 72/138 (52%), Gaps = 1/138 (0%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLDVSF-DFFLERFFYFRIGRRVMVNNLLAM 59
+ +L+ Q H V LL L++ ++ D +FL++ R+G R++ + LA+
Sbjct 151 YCQLVRQLLDDHKDVVTLLAEGLRESRKHIEDEKLVRYFLDKTLTSRLGIRMLATHHLAL 210
Query 60 QNPKPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVVAGNLETEFATIPEHLA 119
KP + GI+ R P ++I R C+ YG AP V + G++ F IP L
Sbjct 211 HEDKPDFFGIICTRLSPKKIIEKWVDFARRLCEHKYGNAPRVRINGHVAARFPFIPMPLD 270
Query 120 LIITEVLKNALRATVEFY 137
I+ E+LKNA+RAT+E +
Sbjct 271 YILPELLKNAMRATMESH 288
> Hs4885545
Length=406
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 77/153 (50%), Gaps = 16/153 (10%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLK-RLSLD----VSFDFFLERFFYFRIGRRVMVNN 55
F+++L + R +H+ V + + + + K + D + +FL+RF+ RI R+++N
Sbjct 108 FLQVLIKVRNRHNDVVPTMAQGVIEYKEKFGFDPFISTNIQYFLDRFYTNRISFRMLINQ 167
Query 56 LLAM----QNP-KPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVV------A 104
+ NP P G ++P C A+V+ + + C+ Y +AP + V A
Sbjct 168 HTLLFGGDTNPVHPKHIGSIDPTCNVADVVKDAYETAKMLCEQYYLVAPELEVEEFNAKA 227
Query 105 GNLETEFATIPEHLALIITEVLKNALRATVEFY 137
+ + +P HL ++ E+ KN++RATVE Y
Sbjct 228 PDKPIQVVYVPSHLFHMLFELFKNSMRATVELY 260
> 7303893
Length=413
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 41/159 (25%), Positives = 72/159 (45%), Gaps = 18/159 (11%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKR-------LSLDVSFDFFLERFFYFRIGRRVMV 53
FV L+ R +H+ V + + + ++K + S +FL+R + RI R+++
Sbjct 112 FVADLDLIRNRHNDVVQTMAQGVIEMKENEGGQVDAPTESSIQYFLDRLYMSRISIRMLI 171
Query 54 N--NLLAMQNPKPG--WCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVV------ 103
N LL NP G G ++P C ++V+ + R C Y +P + +
Sbjct 172 NQHTLLFGGNPHAGGRHIGCLDPACDLSDVVRDAYENARFLCDQYYLTSPALEIQQHSSE 231
Query 104 -AGNLETEFATIPEHLALIITEVLKNALRATVEFYTLGN 141
NL +P HL ++ E+ KN++RA VE + N
Sbjct 232 PGDNLPIRTVYVPSHLYYMLFELFKNSMRAVVEHHGHDN 270
> Hs4505693
Length=411
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 40/150 (26%), Positives = 68/150 (45%), Gaps = 15/150 (10%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSL-----DVSFDFFLERFFYFRIGRRVMVNN 55
FV L + R +H + V + + + + K + + +FL+RF+ RI R+++N
Sbjct 116 FVDTLIKVRNRHHNVVPTMAQGIIEYKDACTVDPVTNQNLQYFLDRFYMNRISTRMLMNQ 175
Query 56 LLAM----QNPKPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAP---LVVVAGNLE 108
+ + Q P G ++P C V+ + R C Y +P L V G
Sbjct 176 HILIFSDSQTGNPSHIGSIDPNCDVVAVVQDAFECSRMLCDQYYLSSPELKLTQVNGKFP 235
Query 109 TE---FATIPEHLALIITEVLKNALRATVE 135
+ +P HL ++ E+ KNA+RATVE
Sbjct 236 DQPIHIVYVPSHLHHMLFELFKNAMRATVE 265
> Hs19923736
Length=407
Score = 51.6 bits (122), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 40/151 (26%), Positives = 67/151 (44%), Gaps = 16/151 (10%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLD-----VSFDFFLERFFYFRIGRRVMVNN 55
F L R +H+ V + + + + K D + +FL+RF+ RI R+++N
Sbjct 112 FTDALVTIRNRHNDVVPTMAQGVLEYKDTYGDDPVSNQNIQYFLDRFYLSRISIRMLINQ 171
Query 56 ----LLAMQNP-KPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVV----AGN 106
NP P G ++P C +EV+ + C Y +P + + A N
Sbjct 172 HTLIFDGSTNPAHPKHIGSIDPNCNVSEVVKDAYDMAKLLCDKYYMASPDLEIQEINAAN 231
Query 107 LET--EFATIPEHLALIITEVLKNALRATVE 135
+ +P HL ++ E+ KNA+RATVE
Sbjct 232 SKQPIHMVYVPSHLYHMLFELFKNAMRATVE 262
> CE00397
Length=401
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/147 (24%), Positives = 67/147 (45%), Gaps = 15/147 (10%)
Query 10 QQHDSSVDLLGRALQQLKR-----LSLDVSFDFFLERFFYFRIGRRVMVN-NLLAMQN-- 61
++H V+ + L +L+ ++ + +FL+RF+ RI R++ N +L+ N
Sbjct 118 KRHAHVVETMAEGLIELRESDGVDIASEKGIQYFLDRFYINRISIRMLQNQHLVVFGNVL 177
Query 62 -PKPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLV------VVAGNLETEFATI 114
P G ++P C V+ + R C Y +P + V +
Sbjct 178 PESPRHVGCIDPACDVESVVYDAFENARFLCDRYYLTSPSMKLEMHNAVEKGKPISIVAV 237
Query 115 PEHLALIITEVLKNALRATVEFYTLGN 141
P HL ++ E+ KNA+RATVE++ + +
Sbjct 238 PSHLYHMMFELFKNAMRATVEYHGVDD 264
> Hs4505689
Length=436
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 34/115 (29%), Positives = 53/115 (46%), Gaps = 13/115 (11%)
Query 37 FFLERFFYFRIGRRVMVN--NLLAMQNPKPG-----WCGIVNPRCRPAEVIMARAQEVRD 89
+FL+RF+ RI R+++N +LL K G +NP C EVI + R
Sbjct 179 YFLDRFYMSRISIRMLLNQHSLLFGGKGKGSPSHRKHIGSINPNCNVLEVIKDGYENARR 238
Query 90 SCKFSYGIAP------LVVVAGNLETEFATIPEHLALIITEVLKNALRATVEFYT 138
C Y +P L + + +P HL ++ E+ KNA+RAT+E +
Sbjct 239 LCDLYYINSPELELEELNAKSPGQPIQVVYVPSHLYHMVFELFKNAMRATMEHHA 293
> YIL042c
Length=394
Score = 45.4 bits (106), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 36/141 (25%), Positives = 65/141 (46%), Gaps = 11/141 (7%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLDVSFDFFLERFFYFRIGRRVMVNNLLAMQ 60
F +LL+ H+ ++ +L + LQ+++ FL RI +++V + L++
Sbjct 141 FTELLDD----HEDAIVVLAKGLQEIQSCYPKFQISQFLNFHLKERITMKLLVTHYLSLM 196
Query 61 NPKPG-----WCGIVNPRCRPAEVIMARAQEVRDSC--KFSYGIAPLVVVAGNLETEFAT 113
G GI++ A++I + V D C KF+ P+++ + + F
Sbjct 197 AQNKGDTNKRMIGILHRDLPIAQLIKHVSDYVNDICFVKFNTQRTPVLIHPPSQDITFTC 256
Query 114 IPEHLALIITEVLKNALRATV 134
IP L I+TEV KNA A +
Sbjct 257 IPPILEYIMTEVFKNAFEAQI 277
> YGL059w
Length=445
Score = 33.1 bits (74), Expect = 0.19, Method: Composition-based stats.
Identities = 12/39 (30%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 99 PLVVVAGNLETEFATIPEHLALIITEVLKNALRATVEFY 137
P ++ G+ + F +P HL ++ E+L+N AT++ Y
Sbjct 275 PEFIIEGDTQLSFYFLPTHLKYLLGEILRNTYEATMKHY 313
> 7295968
Length=1039
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 44 YFRIGRRVMVNNLLAMQNPKPGWCGIVNPRCRPAEVIM 81
Y ++VN+ LAM +P W ++ PR E I+
Sbjct 14 YTHCNASLLVNDTLAMSGDQPTWLKLLPPRLHTPEAIL 51
> At4g23620
Length=264
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 4 LLEQERQQHDSSVDLLGRALQQLKRLSLDVSFDFFLERFF 43
+ EQE QH + L+ Q+++L + + FFL R F
Sbjct 80 IFEQEDGQHGGNKRLISVQTNQIRKLVNHLGYSFFLSRLF 119
> 7289585
Length=384
Score = 28.5 bits (62), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 30/68 (44%), Gaps = 9/68 (13%)
Query 76 PAEVIMARAQEVRDSCKFSYGIAPLVVVAGNLETEFATIPEHLALIITEV--LKNALRAT 133
P +V EV+D C YG V++ F T+PE I E +++A++
Sbjct 319 PGDVDEELEPEVKDECNTKYGEVNSVII----HESFGTVPEDAVKIFVEFRRIESAIKG- 373
Query 134 VEFYTLGN 141
FYT N
Sbjct 374 --FYTYVN 379
> At3g50100
Length=406
Score = 28.1 bits (61), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 4/64 (6%)
Query 1 FVKLLEQERQQHDSSVDLLGRALQQLKRLSLDVSFDFFLERFFYFRIG---RRVM-VNNL 56
++ L+QE Q D+ +L R + SLD SF + E +F +G ++VM N+
Sbjct 81 LIENLKQESQDEDTPEQMLVRLTVEHPSYSLDYSFKPYSEDWFVSDVGMKMKKVMESTNM 140
Query 57 LAMQ 60
+A+
Sbjct 141 VAVD 144
> YKL080w
Length=392
Score = 27.7 bits (60), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 29/122 (23%), Positives = 57/122 (46%), Gaps = 5/122 (4%)
Query 3 KLLEQERQQHDSSVDLLGRALQQLKRLSLDVSFDFFLERFFYFRIGRRVMVNNLLAMQNP 62
+L++Q +++HDS+ L QL RL+ D F+ +F+ + RV V ++L P
Sbjct 265 ELIDQLKKEHDSAASLEQSLRVQLVRLAKTAYVDVFI-NWFHIK-ALRVYVESVLRYGLP 322
Query 63 KPGWCGIVNPRCRPAEVIMARAQEVRDSCKFSYGIAPLVVVAGNLETEFATIPEHLALII 122
I+ P + + E+ D+ F G A + G + + ++ ++ +L+
Sbjct 323 PHFNIKII---AVPPKNLSKCKSELIDAFGFLGGNAFMKDKKGKINKQDTSLHQYASLVD 379
Query 123 TE 124
TE
Sbjct 380 TE 381
Lambda K H
0.327 0.141 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1608078824
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40