bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3249_orf2
Length=171
Score E
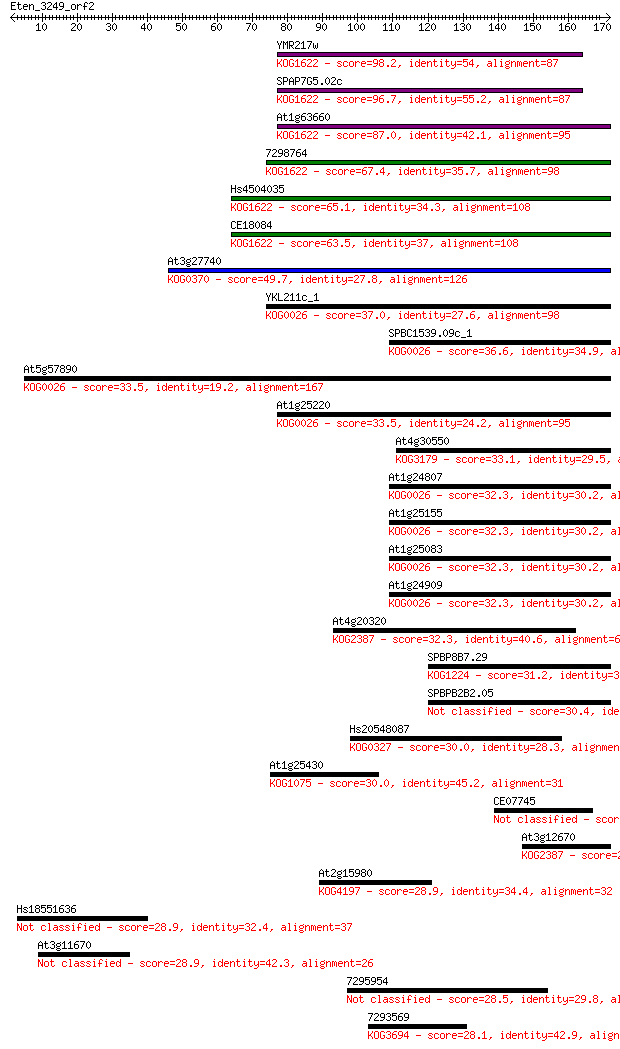
Sequences producing significant alignments: (Bits) Value
YMR217w 98.2 6e-21
SPAP7G5.02c 96.7 2e-20
At1g63660 87.0 2e-17
7298764 67.4 1e-11
Hs4504035 65.1 7e-11
CE18084 63.5 2e-10
At3g27740 49.7 3e-06
YKL211c_1 37.0 0.020
SPBC1539.09c_1 36.6 0.028
At5g57890 33.5 0.20
At1g25220 33.5 0.22
At4g30550 33.1 0.31
At1g24807 32.3 0.42
At1g25155 32.3 0.43
At1g25083 32.3 0.43
At1g24909 32.3 0.43
At4g20320 32.3 0.47
SPBP8B7.29 31.2 0.92
SPBPB2B2.05 30.4 1.8
Hs20548087 30.0 2.5
At1g25430 30.0 2.6
CE07745 29.6 3.0
At3g12670 29.3 4.0
At2g15980 28.9 4.8
Hs18551636 28.9 5.0
At3g11670 28.9 5.7
7295954 28.5 6.4
7293569 28.1 9.0
> YMR217w
Length=525
Score = 98.2 bits (243), Expect = 6e-21, Method: Composition-based stats.
Identities = 47/87 (54%), Positives = 58/87 (66%), Gaps = 5/87 (5%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
+LV DFGSQ S LI RRLR +YAE+L CT + E+ P V+LSGGP SVY E AP
Sbjct 14 ILVLDFGSQYSHLITRRLREFNIYAEMLPCTQKISELGWT-PKGVILSGGPYSVYAEDAP 72
Query 137 HLRREVWQLLQQQQIPILGICYGMQEI 163
H+ ++ L +PILGICYGMQE+
Sbjct 73 HVDHAIFDL----NVPILGICYGMQEL 95
> SPAP7G5.02c
Length=539
Score = 96.7 bits (239), Expect = 2e-20, Method: Composition-based stats.
Identities = 48/87 (55%), Positives = 62/87 (71%), Gaps = 5/87 (5%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
+L+ DFGSQ S LI RRLR + VYAELL CT +E + +P V+LSGGP SVY++ AP
Sbjct 21 ILILDFGSQYSHLIARRLREIHVYAELLPCTQKIEAL-PFKPIGVILSGGPYSVYDDIAP 79
Query 137 HLRREVWQLLQQQQIPILGICYGMQEI 163
H+ V++L +P+LGICYGMQEI
Sbjct 80 HVDPAVFEL----GVPVLGICYGMQEI 102
> At1g63660
Length=534
Score = 87.0 bits (214), Expect = 2e-17, Method: Composition-based stats.
Identities = 40/95 (42%), Positives = 63/95 (66%), Gaps = 0/95 (0%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAP 136
VL+ D+GSQ + LI RR+R++ V++ ++S T++L+ + + P V+LSGGP SV+ AP
Sbjct 11 VLILDYGSQYTHLITRRIRSLNVFSLVISGTSSLKSITSYNPRVVILSGGPHSVHALDAP 70
Query 137 HLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + + +LGICYG+Q IVQ+LGG V
Sbjct 71 SFPEGFIEWAESNGVSVLGICYGLQLIVQKLGGVV 105
> 7298764
Length=683
Score = 67.4 bits (163), Expect = 1e-11, Method: Composition-based stats.
Identities = 35/99 (35%), Positives = 60/99 (60%), Gaps = 5/99 (5%)
Query 74 RH-MVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYE 132
RH +++ D G+Q ++I R++R + V ++L + +++SGGP+SVY
Sbjct 15 RHDKIVILDAGAQYGKVIDRKVRELFVETDILPLDTPAATIRNNGYRGIIISGGPNSVYA 74
Query 133 EGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
E AP ++++L +IP+LGICYGMQ I ++ GG+V
Sbjct 75 EDAPSYDPDLFKL----KIPMLGICYGMQLINKEFGGTV 109
> Hs4504035
Length=693
Score = 65.1 bits (157), Expect = 7e-11, Method: Composition-based stats.
Identities = 37/112 (33%), Positives = 61/112 (54%), Gaps = 8/112 (7%)
Query 64 ELSGGRRRPARH----MVLVFDFGSQVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPA 119
E +GG + H V++ D G+Q ++I RR+R + V +E+ +
Sbjct 11 ENAGGDLKDGHHHYEGAVVILDAGAQYGKVIDRRVRELFVQSEIFPLETPAFAIKEQGFR 70
Query 120 AVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
A+++SGGP+SVY E AP ++ + + P+LGICYGMQ + + GG+V
Sbjct 71 AIIISGGPNSVYAEDAPWFDPAIFTIGK----PVLGICYGMQMMNKVFGGTV 118
> CE18084
Length=792
Score = 63.5 bits (153), Expect = 2e-10, Method: Composition-based stats.
Identities = 40/111 (36%), Positives = 67/111 (60%), Gaps = 15/111 (13%)
Query 64 ELSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAEL--LSCTA-TLEEVAAARPAA 120
++S G R + + DFG+Q ++I RR+R + V +E+ L+ TA TL E+ +
Sbjct 101 KVSSGER------IAILDFGAQYGKVIDRRVRELLVQSEMFPLNTTARTLIELGGFK--G 152
Query 121 VVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+++SGGP+SV+E AP + E++ +P+LGICYG Q + + GG+V
Sbjct 153 IIISGGPNSVFEPEAPSIDPEIFTC----GLPVLGICYGFQLMNKLNGGTV 199
> At3g27740
Length=428
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 35/126 (27%), Positives = 54/126 (42%), Gaps = 9/126 (7%)
Query 46 CAFLREIVSKMTVYSRESELSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAELLS 105
C E V K E + + R + V+ +DFG + + I+RRL + G ++
Sbjct 214 CKSPYEWVDKTNA---EWDFNTNSRDGKSYKVIAYDFG--IKQNILRRLSSYGCQITVVP 268
Query 106 CTATLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQ 165
T E P ++ S GP P+ V +LL + +P+ GIC G Q + Q
Sbjct 269 STFPAAEALKMNPDGILFSNGPGD--PSAVPYAVETVKELLGK--VPVYGICMGHQLLGQ 324
Query 166 QLGGSV 171
LGG
Sbjct 325 ALGGKT 330
> YKL211c_1
Length=206
Score = 37.0 bits (84), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 47/101 (46%), Gaps = 9/101 (8%)
Query 74 RHMVLVFDFGS---QVSRLIIRRLRAVGVYAELLSCTATLEEVAAARPAAVVLSGGPSSV 130
+H+VL+ ++ S V + + V VY + T+ E+AA P +++S GP
Sbjct 12 KHVVLIDNYDSFTWNVYEYLCQEGAKVSVYR---NDAITVPEIAALNPDTLLISPGPGHP 68
Query 131 YEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ + R+ + + IP+ GIC G Q + GG V
Sbjct 69 KTDSG--ISRDCIRYFTGK-IPVFGICMGQQCMFDVFGGEV 106
> SPBC1539.09c_1
Length=238
Score = 36.6 bits (83), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 33/63 (52%), Gaps = 3/63 (4%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T++E+ P +VLS GP +G + E + IPILG+C G+Q I + +G
Sbjct 63 TVDELEKLNPLKLVLSPGPGHPARDGG--ICNEAISRFAGK-IPILGVCMGLQCIFETMG 119
Query 169 GSV 171
G V
Sbjct 120 GKV 122
> At5g57890
Length=273
Score = 33.5 bits (75), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 32/168 (19%), Positives = 76/168 (45%), Gaps = 9/168 (5%)
Query 5 SSLLSICKVKSEYLWLSPAAEAPHIEQEKNPQGSGVIFFPFCAFLREIVSKMTVYSRESE 64
++L + C ++ +Y + + + NP V++ R++++K ++ ES
Sbjct 4 TTLYNSCLLQPKYGFTTRRLNQSLVNSLTNPTRVSVLW----KSRRDVIAKASIEMAESN 59
Query 65 LSGGRRRPARHMVLVFDFGSQVSRLIIRRLRAVGVYAELL-SCTATLEEVAAARPAAVVL 123
+ ++V D + + + + +G + E+ + T+EE+ +P +++
Sbjct 60 SISSVVVNSSGPIIVIDNYDSFTYNLCQYMGELGCHFEVYRNDELTVEELKRKKPRGLLI 119
Query 124 SGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
S GP + + G + V +L +P+ G+C G+Q I + GG +
Sbjct 120 SPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFGGKI 163
> At1g25220
Length=276
Score = 33.5 bits (75), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 47/96 (48%), Gaps = 5/96 (5%)
Query 77 VLVFDFGSQVSRLIIRRLRAVGVYAELL-SCTATLEEVAAARPAAVVLSGGPSSVYEEGA 135
++V D + + + + +G + E+ + T+EE+ P V++S GP + + G
Sbjct 75 IIVIDNYDSFTYNLCQYMGELGCHFEVYRNDELTVEELKKKNPRGVLISPGPGTPQDSGI 134
Query 136 PHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
+ V +L +P+ G+C G+Q I + GG +
Sbjct 135 S--LQTVLEL--GPLVPLFGVCMGLQCIGEAFGGKI 166
> At4g30550
Length=249
Score = 33.1 bits (74), Expect = 0.31, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 30/62 (48%), Gaps = 1/62 (1%)
Query 111 EEVAAARPAAVVLSGGPSSVYEEGAPHLRR-EVWQLLQQQQIPILGICYGMQEIVQQLGG 169
+E + V+SG P + + ++ EV Q L + +LGIC+G Q I + GG
Sbjct 56 DENDLDKYDGFVISGSPHDAFGDADWIVKLCEVCQKLDHMKKKVLGICFGHQIITRVKGG 115
Query 170 SV 171
+
Sbjct 116 KI 117
> At1g24807
Length=235
Score = 32.3 bits (72), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 67 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 122
Query 169 GSV 171
G +
Sbjct 123 GKI 125
> At1g25155
Length=222
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> At1g25083
Length=222
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> At1g24909
Length=222
Score = 32.3 bits (72), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 4/63 (6%)
Query 109 TLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLG 168
T+EE+ P V++S GP + + G + V +L +P+ G+C G+Q I + G
Sbjct 54 TVEELKRKNPRGVLISPGPGTPQDSGIS--LQTVLEL--GPLVPLFGVCMGLQCIGEAFG 109
Query 169 GSV 171
G +
Sbjct 110 GKI 112
> At4g20320
Length=553
Score = 32.3 bits (72), Expect = 0.47, Method: Composition-based stats.
Identities = 28/104 (26%), Positives = 40/104 (38%), Gaps = 35/104 (33%)
Query 93 RLRAVGVYAELL----SCTATLEEVAAARPAAVVLSGGPSSVYEEGA----PHLRREVWQ 144
R+ VG Y ELL S L + AR +++ +S E+GA P + W+
Sbjct 289 RIAVVGKYTELLDSYLSIHKALLHASVARRKKLIIDWISASDLEQGAKKENPDAYKAAWK 348
Query 145 LLQ---------------------------QQQIPILGICYGMQ 161
LL+ + +IP LGIC GMQ
Sbjct 349 LLKGADGVLVPGGFGSRGVEGKMLAAKYARENRIPYLGICLGMQ 392
> SPBP8B7.29
Length=718
Score = 31.2 bits (69), Expect = 0.92, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 26/52 (50%), Gaps = 6/52 (11%)
Query 120 AVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGICYGMQEIVQQLGGSV 171
A+V+ GP E + + +WQL IP++GIC G Q + G ++
Sbjct 57 AIVVGPGPGHPAEYSS--ILNRIWQL----NIPVMGICLGFQSLALYHGATI 102
> SPBPB2B2.05
Length=237
Score = 30.4 bits (67), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 15/67 (22%)
Query 120 AVVLSGG----PSSVYEEGAPHLRREV--------WQLLQ---QQQIPILGICYGMQEIV 164
++L+GG P+ E+ P+ + V W ++ +++IPILGIC G Q +
Sbjct 32 GIILAGGESVHPNRYGEDFDPNAPKSVDVIRDSTEWGMIDFALKKKIPILGICRGCQVLN 91
Query 165 QQLGGSV 171
GGS+
Sbjct 92 VYFGGSL 98
> Hs20548087
Length=344
Score = 30.0 bits (66), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 10/60 (16%)
Query 98 GVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPILGIC 157
G++ EL S T + P+A + PS V E +R +W L++++++ + GIC
Sbjct 232 GIFQELNSNTQVV------LPSATM----PSDVLEVTKKFMREHIWILVKKEELTLEGIC 281
> At1g25430
Length=1213
Score = 30.0 bits (66), Expect = 2.6, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 4/35 (11%)
Query 75 HMVLVFDFGSQVSRLIIRRL----RAVGVYAELLS 105
H++L+ +F +QV RL+ RR+ R ++ELLS
Sbjct 1105 HLLLICEFSAQVWRLVFRRICPRQRLFSSWSELLS 1139
> CE07745
Length=471
Score = 29.6 bits (65), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 21/31 (67%), Gaps = 3/31 (9%)
Query 139 RREVWQLLQQQQI-PIL--GICYGMQEIVQQ 166
+ + WQLL+ ++I P L GI YG QE VQQ
Sbjct 402 KDQYWQLLKDERIIPFLAYGISYGDQETVQQ 432
> At3g12670
Length=591
Score = 29.3 bits (64), Expect = 4.0, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 147 QQQQIPILGICYGMQEIVQQLGGSV 171
++ Q+P LGIC GMQ V + S+
Sbjct 387 RENQVPFLGICLGMQLAVVEFARSI 411
> At2g15980
Length=498
Score = 28.9 bits (63), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 11/32 (34%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 89 LIIRRLRAVGVYAELLSCTATLEEVAAARPAA 120
+++R+LR+ G+ A++ +C A + EV+ R A+
Sbjct 183 MVMRKLRSRGINAQISTCNALITEVSRRRGAS 214
> Hs18551636
Length=1539
Score = 28.9 bits (63), Expect = 5.0, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 23/37 (62%), Gaps = 3/37 (8%)
Query 3 QLSSLLSICKVKSEYLWLSPAAEAPHIEQEKNPQGSG 39
+L+S S C ++ + L P+ + P +++E+ PQG G
Sbjct 312 KLTSCASSCPLEMK---LCPSVQTPQVQRERGPQGQG 345
> At3g11670
Length=808
Score = 28.9 bits (63), Expect = 5.7, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 9 SICKVKSEYLWLSPAAEAPHIEQEKN 34
S+CK KS+ LW P+A+A + + N
Sbjct 281 SLCKCKSQQLWRLPSAQASDLIENDN 306
> 7295954
Length=487
Score = 28.5 bits (62), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query 97 VGVYAELLSCTATLEEVAAARPAAVVLSGGPSSVYEEGAPHLRREVWQLLQQQQIPI 153
+G EL++C T++ AAA P+ ++ V+EE +P +RR Q +Q+ +
Sbjct 219 IGDPLELIACGPTIQPEAAASPSDILKK---HHVWEELSPEIRRVFEQPEEQKNTSL 272
> 7293569
Length=1922
Score = 28.1 bits (61), Expect = 9.0, Method: Composition-based stats.
Identities = 12/28 (42%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 103 LLSCTATLEEVAAARPAAVVLSGGPSSV 130
L +C ATL E A PA +++ GG +S+
Sbjct 175 LRACLATLAEFAVLNPALLIVCGGVTSI 202
Lambda K H
0.322 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2526689620
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40