bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3269_orf1
Length=346
Score E
Sequences producing significant alignments: (Bits) Value
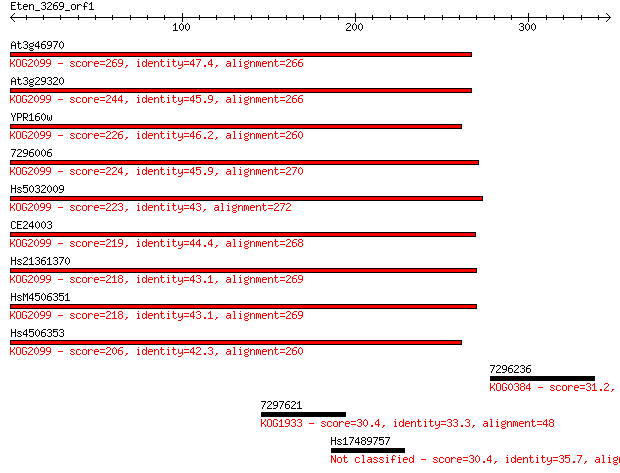
At3g46970 269 7e-72
At3g29320 244 2e-64
YPR160w 226 5e-59
7296006 224 2e-58
Hs5032009 223 6e-58
CE24003 219 5e-57
Hs21361370 218 1e-56
HsM4506351 218 2e-56
Hs4506353 206 4e-53
7296236 31.2 3.7
7297621 30.4 5.2
Hs17489757 30.4 5.5
> At3g46970
Length=841
Score = 269 bits (687), Expect = 7e-72, Method: Compositional matrix adjust.
Identities = 126/266 (47%), Positives = 180/266 (67%), Gaps = 6/266 (2%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
+NI+ +++R+ LK M+P +R +PR +IGGKA Y AK I+K+ N+V +VN+DP
Sbjct 582 MNILGVVYRFKKLKEMKPEERKKTVPRTVMIGGKAFATYTNAKRIVKLVNDVGDVVNSDP 641
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
+V++YLKVVF+PNYNV+ A+++IP S++SQHISTAG EASGTSNMKF +NG LIIGTLDG
Sbjct 642 EVNEYLKVVFVPNYNVTVAEMLIPGSELSQHISTAGMEASGTSNMKFALNGCLIIGTLDG 701
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDHRMREVFDWIRSGALACGDDK 180
AN+EIREE G+E F+FGA +V +R+ +G + D R E +++SG
Sbjct 702 ANVEIREEVGEENFFLFGATADQVPRLRKEREDGLFKPDPRFEEAKQFVKSGVFGS---- 757
Query 181 SHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAAAS 240
D+ +LD++ N GDY+L+ +DFP Y AQ +VD YK+ W ++SI + A
Sbjct 758 --YDYGPLLDSLEGNTGFGRGDYFLVGYDFPSYMDAQAKVDEAYKDRKGWLKMSILSTAG 815
Query 241 MGKFSTDRCMREYAEKIWGISRCERP 266
GKFS+DR + +YA++IW I C P
Sbjct 816 SGKFSSDRTIAQYAKEIWNIEACPVP 841
> At3g29320
Length=962
Score = 244 bits (623), Expect = 2e-64, Method: Compositional matrix adjust.
Identities = 122/267 (45%), Positives = 172/267 (64%), Gaps = 7/267 (2%)
Query 1 LNIIYIIHRYLTLKRMQPADRSN-MLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNND 59
LNI+ I++RY +K M ++R +PR C+ GGKA Y AK I+K +VA +N+D
Sbjct 702 LNILGIVYRYKKMKEMSASEREKAFVPRVCIFGGKAFATYVQAKRIVKFITDVASTINHD 761
Query 60 PDVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLD 119
P++ LKV+F+P+YNVS A+++IPAS++SQHISTAG EASGTSNMKF MNG ++IGTLD
Sbjct 762 PEIGDLLKVIFVPDYNVSVAELLIPASELSQHISTAGMEASGTSNMKFSMNGCVLIGTLD 821
Query 120 GANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDHRMREVFDWIRSGALACGDD 179
GAN+EIREE G+E F+FGAK ++ ++R+ G + D EV ++ SG
Sbjct 822 GANVEIREEVGEENFFLFGAKADQIVNLRKERAEGKFVPDPTFEEVKKFVGSGVFGSN-- 879
Query 180 KSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAAA 239
S+ + L+ N G G DY+L+ DFP Y Q++VD Y++ +W R+SI A
Sbjct 880 -SYDELIGSLEG--NEGFG-RADYFLVGKDFPSYIECQEKVDEAYRDQKRWTRMSIMNTA 935
Query 240 SMGKFSTDRCMREYAEKIWGISRCERP 266
KFS+DR + EYA+ IW I + E P
Sbjct 936 GSFKFSSDRTIHEYAKDIWNIKQVELP 962
> YPR160w
Length=902
Score = 226 bits (576), Expect = 5e-59, Method: Compositional matrix adjust.
Identities = 120/267 (44%), Positives = 171/267 (64%), Gaps = 15/267 (5%)
Query 1 LNIIYIIHRYLTLKRMQP-----ADRSNMLPR-ACLIGGKAAPGYYTAKTIIKMANNVAQ 54
LN+ II+RYL +K M + + PR + GGK+APGYY AK IIK+ N VA
Sbjct 640 LNVFGIIYRYLAMKNMLKNGASIEEVAKKYPRKVSIFGGKSAPGYYMAKLIIKLINCVAD 699
Query 55 IVNNDPDVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLI 114
IVNND ++ LKVVF+ +YNVS A++IIPASD+S+HISTAGTEASGTSNMKFVMNGGLI
Sbjct 700 IVNNDESIEHLLKVVFVADYNVSKAEIIIPASDLSEHISTAGTEASGTSNMKFVMNGGLI 759
Query 115 IGTLDGANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDHRMREVFDWIRSGAL 174
IGT+DGAN+EI E G++ +F+FG V+ +R + + + V +I SG
Sbjct 760 IGTVDGANVEITREIGEDNVFLFGNLSENVEELRYNHQYHPQDLPSSLDSVLSYIESGQF 819
Query 175 ACGDDKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNP-DKWWRL 233
+ ++ +F ++D+I +GDYYL+ DF Y + VD+ + N +W +
Sbjct 820 SP---ENPNEFKPLVDSI-----KYHGDYYLVSDDFESYLATHELVDQEFHNQRSEWLKK 871
Query 234 SIKAAASMGKFSTDRCMREYAEKIWGI 260
S+ + A++G FS+DRC+ EY++ IW +
Sbjct 872 SVLSVANVGFFSSDRCIEEYSDTIWNV 898
> 7296006
Length=844
Score = 224 bits (571), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 124/273 (45%), Positives = 170/273 (62%), Gaps = 15/273 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LN ++II Y R++ +N PR +IGGKAAPGYY AK IIK+ V +VNNDP
Sbjct 579 LNCLHIITLY---NRIKKDPTANFTPRTIMIGGKAAPGYYVAKQIIKLICAVGNVVNNDP 635
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V LKV+FL NY V+ A+ I+PA+D+S+ ISTAGTEASGT NMKF +NG L IGTLDG
Sbjct 636 IVGDKLKVIFLENYRVTLAEKIMPAADLSEQISTAGTEASGTGNMKFQLNGALTIGTLDG 695
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNG--SYPMDHRMREVFDWIRSGALACGD 178
AN+E+ EE G + +FIFG EV+ ++++ N Y + +++V D I+ G + G+
Sbjct 696 ANVEMAEEMGLDNIFIFGMTVDEVEALKKKGYNAYDYYNANPEVKQVIDQIQGGFFSPGN 755
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
+F I D + D+Y L+ D+ Y +AQD V +TY+N KW +SI
Sbjct 756 P---NEFKNIADILLKY------DHYYLLADYDAYIKAQDLVSKTYQNQAKWLEMSINNI 806
Query 239 ASMGKFSTDRCMREYAEKIWGIS-RCERPPPDE 270
AS GKFS+DR + EYA +IWG+ E+ P E
Sbjct 807 ASSGKFSSDRTIAEYAREIWGVEPTWEKLPAPE 839
> Hs5032009
Length=842
Score = 223 bits (567), Expect = 6e-58, Method: Compositional matrix adjust.
Identities = 117/276 (42%), Positives = 170/276 (61%), Gaps = 16/276 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LN +++I Y +KR +P +PR +IGGKAAPGY+ AK II++ + +VN+DP
Sbjct 579 LNCLHVITLYNRIKR-EP--NKFFVPRTVMIGGKAAPGYHMAKMIIRLVTAIGDVVNHDP 635
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V L+V+FL NY VS A+ +IPA+D+S+ ISTAGTEASGT NMKF++NG L IGT+DG
Sbjct 636 AVGDRLRVIFLENYRVSLAEKVIPAADLSEQISTAGTEASGTGNMKFMLNGALTIGTMDG 695
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDH--RMREVFDWIRSGALACGD 178
AN+E+ EE G+E FIFG + +VD + +R N D +R+V + + SG +
Sbjct 696 ANVEMAEEAGEENFFIFGMRVEDVDKLDQRGYNAQEYYDRIPELRQVIEQLSSGFFSPKQ 755
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
D +L + D + + D+ DY + Q++V YKNP +W R+ I+
Sbjct 756 PDLFKDIVNML---------MHHDRFKVFADYEDYIKCQEKVSALYKNPREWTRMVIRNI 806
Query 239 ASMGKFSTDRCMREYAEKIWGI--SRCERPPPDEVL 272
A+ GKFS+DR + +YA +IWG+ SR P PDE +
Sbjct 807 ATSGKFSSDRTIAQYAREIWGVEPSRQRLPAPDEAI 842
> CE24003
Length=882
Score = 219 bits (559), Expect = 5e-57, Method: Compositional matrix adjust.
Identities = 119/270 (44%), Positives = 165/270 (61%), Gaps = 14/270 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LNI+++I Y R++ +M+ R L GGKAAPGY+ AK II++ VA+ VNND
Sbjct 616 LNILHVIALY---NRIKENPNIDMVKRTVLYGGKAAPGYHMAKQIIRLITAVAEQVNNDA 672
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V LKV+FL NY VS A+ IIPASD+S+ ISTAGTEASGT NMKF++NG L IGTLDG
Sbjct 673 IVGDRLKVIFLENYRVSMAEKIIPASDLSEQISTAGTEASGTGNMKFMLNGALTIGTLDG 732
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDH--RMREVFDWIRSGALACGD 178
AN+E+ EE GDE +FIFG EV+ + +R + +D ++++ + I G D
Sbjct 733 ANVEMAEEMGDENIFIFGMNVEEVEALTKRGYSSQEFIDKSPMLKQIIEQIEGGMFTPED 792
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
D +L D +++ DF + AQD+V T+++ +KW R+++
Sbjct 793 PDQLKDLSNML---------RYHDRFMVCADFDAFIEAQDKVSATFRDQEKWSRMALYNI 843
Query 239 ASMGKFSTDRCMREYAEKIWGISRCERPPP 268
AS GKFSTDR + EYA +IWGI + E P
Sbjct 844 ASTGKFSTDRTITEYAREIWGIDQFESSLP 873
> Hs21361370
Length=843
Score = 218 bits (555), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 116/273 (42%), Positives = 168/273 (61%), Gaps = 16/273 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LN ++++ Y +KR PA +PR +IGGKAAPGY+ AK IIK+ ++ +VN+DP
Sbjct 579 LNCLHVVTLYNRIKR-DPA--KAFVPRTVMIGGKAAPGYHMAKLIIKLVTSIGDVVNHDP 635
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V LKV+FL NY VS A+ +IPA+D+SQ ISTAGTEASGT NMKF++NG L IGT+DG
Sbjct 636 VVGDRLKVIFLENYRVSLAEKVIPAADLSQQISTAGTEASGTGNMKFMLNGALTIGTMDG 695
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDH--RMREVFDWIRSGALACGD 178
AN+E+ EE G E +FIFG + +V+ + + N DH +++ D I SG + +
Sbjct 696 ANVEMAEEAGAENLFIFGLRVEDVEALDRKGYNAREYYDHLPELKQAVDQISSGFFSPKE 755
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
D +L + D + + D+ Y + Q +VD+ Y+NP +W + I+
Sbjct 756 PDCFKDIVNML---------MHHDRFKVFADYEAYMQCQAQVDQLYRNPKEWTKKVIRNI 806
Query 239 ASMGKFSTDRCMREYAEKIWGI--SRCERPPPD 269
A GKFS+DR + EYA +IWG+ S + PPP+
Sbjct 807 ACSGKFSSDRTITEYAREIWGVEPSDLQIPPPN 839
> HsM4506351
Length=843
Score = 218 bits (555), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 116/273 (42%), Positives = 168/273 (61%), Gaps = 16/273 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LN ++++ Y +KR PA +PR +IGGKAAPGY+ AK IIK+ ++ +VN+DP
Sbjct 579 LNCLHVVTLYNRIKR-DPA--KAFVPRTVMIGGKAAPGYHMAKLIIKLVTSIGDVVNHDP 635
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V LKV+FL NY VS A+ +IPA+D+SQ ISTAGTEASGT NMKF++NG L IGT+DG
Sbjct 636 VVGDRLKVIFLENYRVSLAEKVIPAADLSQQISTAGTEASGTGNMKFMLNGALTIGTMDG 695
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGSYPMDH--RMREVFDWIRSGALACGD 178
AN+E+ EE G E +FIFG + +V+ + + N DH +++ D I SG + +
Sbjct 696 ANVEMAEEAGAENLFIFGLRVEDVEALDRKGYNAREYYDHLPELKQAVDQISSGFFSPKE 755
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
D +L + D + + D+ Y + Q +VD+ Y+NP +W + I+
Sbjct 756 PDCFKDIVNML---------MHHDRFKVFADYEAYMQCQAQVDQLYRNPKEWTKKVIRNI 806
Query 239 ASMGKFSTDRCMREYAEKIWGI--SRCERPPPD 269
A GKFS+DR + EYA +IWG+ S + PPP+
Sbjct 807 ACSGKFSSDRTITEYAREIWGVEPSDLQIPPPN 839
> Hs4506353
Length=847
Score = 206 bits (525), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 110/262 (41%), Positives = 162/262 (61%), Gaps = 14/262 (5%)
Query 1 LNIIYIIHRYLTLKRMQPADRSNMLPRACLIGGKAAPGYYTAKTIIKMANNVAQIVNNDP 60
LN +++I Y R++ + +PR +IGGKAAPGY+ AK IIK+ +VA +VNNDP
Sbjct 579 LNCLHVITMY---NRIKKDPKKLFVPRTVIIGGKAAPGYHMAKMIIKLITSVADVVNNDP 635
Query 61 DVDKYLKVVFLPNYNVSNAQVIIPASDISQHISTAGTEASGTSNMKFVMNGGLIIGTLDG 120
V LKV+FL NY VS A+ +IPA+D+S+ ISTAGTEASGT NMKF++NG L IGT+DG
Sbjct 636 MVGSKLKVIFLENYRVSLAEKVIPATDLSEQISTAGTEASGTGNMKFMLNGALTIGTMDG 695
Query 121 ANIEIREEGGDETMFIFGAKEHEVDHVRERARNGS--YPMDHRMREVFDWIRSGALACGD 178
AN+E+ EE G+E +FIFG + +V + ++ Y ++ V D I +G +
Sbjct 696 ANVEMAEEAGEENLFIFGMRIDDVAALDKKGYEAKEYYEALPELKLVIDQIDNGFFS--- 752
Query 179 DKSHGDFCAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNPDKWWRLSIKAA 238
K F I++ + + D + + D+ Y + QD+V + Y NP W + +K
Sbjct 753 PKQPDLFKDIINMLFYH------DRFKVFADYEAYVKCQDKVSQLYMNPKAWNTMVLKNI 806
Query 239 ASMGKFSTDRCMREYAEKIWGI 260
A+ GKFS+DR ++EYA+ IW +
Sbjct 807 AASGKFSSDRTIKEYAQNIWNV 828
> 7296236
Length=2237
Score = 31.2 bits (69), Expect = 3.7, Method: Composition-based stats.
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 6/62 (9%)
Query 278 ANDKKSNAGPSHGLNRSNNNNSNTTSTNTNSNESSGTDNLRVPGESESQKRGKH--KKEN 335
AND +S + G + + ++SN T N+ S L + GE E K G+ KKEN
Sbjct 1798 ANDDQSAVSKAEGSDEKSTDDSNPEEATTEKNKES----LEIEGEKERVKEGEESVKKEN 1853
Query 336 KQ 337
+
Sbjct 1854 DE 1855
> 7297621
Length=1157
Score = 30.4 bits (67), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 23/51 (45%), Gaps = 3/51 (5%)
Query 146 HVRERARNGSY---PMDHRMREVFDWIRSGALACGDDKSHGDFCAILDTIC 193
++ R N +Y P + + FDW + + C K GDFC DT C
Sbjct 795 YLASRHSNQTYIARPASSWIDDYFDWAAAASSCCKYRKDSGDFCPHQDTSC 845
> Hs17489757
Length=115
Score = 30.4 bits (67), Expect = 5.5, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 22/42 (52%), Gaps = 3/42 (7%)
Query 186 CAILDTICNNGSGNNGDYYLLMHDFPDYCRAQDEVDRTYKNP 227
C + +T CN+G N G Y L DFP Y + E+ ++P
Sbjct 30 CWMPETCCNSGVRNKGHYQL---DFPPYSEREPEICSGVESP 68
Lambda K H
0.315 0.132 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8340552822
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40