bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3272_orf2
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
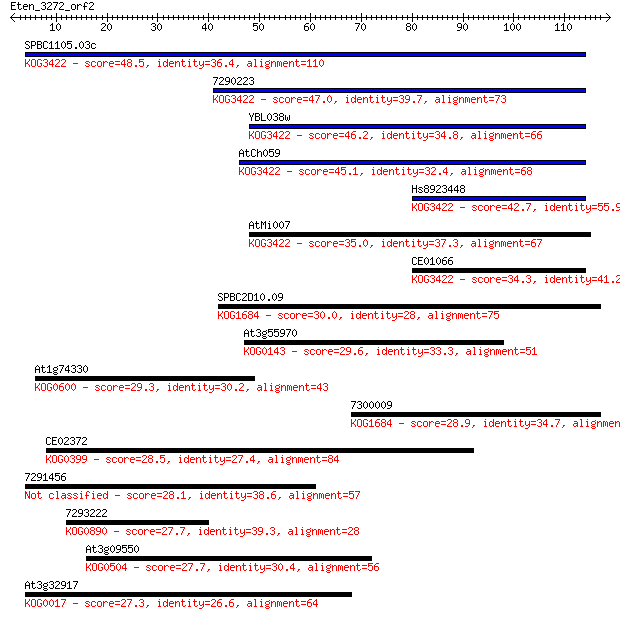
SPBC1105.03c 48.5 3e-06
7290223 47.0 8e-06
YBL038w 46.2 1e-05
AtCh059 45.1 4e-05
Hs8923448 42.7 1e-04
AtMi007 35.0 0.034
CE01066 34.3 0.062
SPBC2D10.09 30.0 0.95
At3g55970 29.6 1.6
At1g74330 29.3 2.0
7300009 28.9 2.3
CE02372 28.5 3.4
7291456 28.1 4.4
7293222 27.7 5.2
At3g09550 27.7 5.8
At3g32917 27.3 7.0
> SPBC1105.03c
Length=215
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 40/127 (31%), Positives = 63/127 (49%), Gaps = 17/127 (13%)
Query 4 GFKMPPVRFK-------APQQVRI-KCPKGRI-VP----LNEPEVKLGKTIVAAAPH--R 48
G +MPP++ AP+ +R K KGR+ +P L +++ G+ + R
Sbjct 24 GLQMPPIQTMVRWGHQYAPRSIRNQKAQKGRVPIPIGGSLRGTQLEWGEYGMRLKDRSIR 83
Query 49 VTEAQLEQARRTLRRYLG--RSKELLVGVHATYPVTRKAEGVKMGQGKGAIDHFVARVPA 106
QLE A + L+R L ++ + PV K +MG+GKGA +++ AR+P
Sbjct 84 FHAKQLETAEQILKRILKPIKAARVYTRFCCNVPVCVKGNETRMGKGKGAFEYWAARIPI 143
Query 107 GRVLFHI 113
GRVLF I
Sbjct 144 GRVLFEI 150
> 7290223
Length=243
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 42/80 (52%), Gaps = 12/80 (15%)
Query 41 IVAAAPHRVTEAQLEQARRTLRRYLGRSKELLVGVHATY-------PVTRKAEGVKMGQG 93
IVA R+ E R T+ R + + + AT+ P+T+K +G +MG G
Sbjct 90 IVATGGGRLRWGHYEMMRLTIGRKMNVNT-----MFATWRVPAPWQPITKKGQGQRMGGG 144
Query 94 KGAIDHFVARVPAGRVLFHI 113
KGAIDH+V + AGRV+ I
Sbjct 145 KGAIDHYVTPIKAGRVIVEI 164
> YBL038w
Length=232
Score = 46.2 bits (108), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 37/68 (54%), Gaps = 2/68 (2%)
Query 48 RVTEAQLEQARRTLRRYLG--RSKELLVGVHATYPVTRKAEGVKMGQGKGAIDHFVARVP 105
R++ QL++A + RY+ + L + V K +MG+GKG DH++ RVP
Sbjct 84 RISAQQLKEADNAIMRYVRPLNNGHLWRRLCTNVAVCIKGNETRMGKGKGGFDHWMVRVP 143
Query 106 AGRVLFHI 113
G++LF I
Sbjct 144 TGKILFEI 151
> AtCh059
Length=135
Score = 45.1 bits (105), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 39/68 (57%), Gaps = 0/68 (0%)
Query 46 PHRVTEAQLEQARRTLRRYLGRSKELLVGVHATYPVTRKAEGVKMGQGKGAIDHFVARVP 105
P +T Q+E RR + R + R ++ V + PVT + +MG GKG+ +++VA V
Sbjct 39 PAWITSRQIEAGRRAMTRNVRRGGKIWVRIFPDKPVTVRPAETRMGSGKGSPEYWVAVVK 98
Query 106 AGRVLFHI 113
G++L+ +
Sbjct 99 PGKILYEM 106
> Hs8923448
Length=251
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/34 (55%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 80 PVTRKAEGVKMGQGKGAIDHFVARVPAGRVLFHI 113
P+TRK+ G +MG GKGAIDH+V V AGR++ +
Sbjct 130 PITRKSVGHRMGGGKGAIDHYVTPVKAGRLVVEM 163
> AtMi007
Length=179
Score = 35.0 bits (79), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 8/75 (10%)
Query 48 RVTEAQLEQARRTL-----RRYLG---RSKELLVGVHATYPVTRKAEGVKMGQGKGAIDH 99
R++ +E ARR + R G R+ ++ V V A P+T K V+MG+GKG
Sbjct 77 RLSYRAIEAARRAIIGHFHRAMSGQFRRNGKIWVRVFADLPITGKPTEVRMGRGKGNPTG 136
Query 100 FVARVPAGRVLFHIP 114
++ARV G++ F +
Sbjct 137 WIARVSTGQIPFEMD 151
> CE01066
Length=219
Score = 34.3 bits (77), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 23/34 (67%), Gaps = 0/34 (0%)
Query 80 PVTRKAEGVKMGQGKGAIDHFVARVPAGRVLFHI 113
P T+KA+G ++G GKG+I +V V A R++ +
Sbjct 107 PRTKKAQGTRLGGGKGSIQKYVTPVRANRIILEV 140
> SPBC2D10.09
Length=429
Score = 30.0 bits (66), Expect = 0.95, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 32/77 (41%), Gaps = 7/77 (9%)
Query 42 VAAAPHRVTEAQLEQARRTLRRYLGRSKELLVGVHATY--PVTRKAEGVKMGQGKGAIDH 99
+ AA + + +L + R + S L ATY PV G+ MG G G H
Sbjct 119 IKAAALSIQDGKLPEVRHAFAQEYRLSHTL-----ATYQKPVVALMNGITMGGGSGLAMH 173
Query 100 FVARVPAGRVLFHIPQT 116
R+ +F +P+T
Sbjct 174 VPFRIACEDTMFAMPET 190
> At3g55970
Length=363
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 47 HRVTEAQLEQARRTLRRYLGRSKELLVGVHATYPVTRKAEGVKMGQGKGAI 97
H ++ ++QA+ T R + EL +HA P T + G ++G KGAI
Sbjct 90 HGMSPQLMDQAKATWREFFNLPMEL-KNMHANSPKTYEGYGSRLGVEKGAI 139
> At1g74330
Length=445
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 6 KMPPVRFKAPQQVRIKCPKGRIVPLNEPEVKLGKTIVAAAPHR 48
K+P PQQ C + + L+E E+ L +T+++ PH+
Sbjct 352 KLPHAMLFKPQQTYDSCLRETLKDLSETEINLIETLLSIDPHK 394
> 7300009
Length=351
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Query 68 SKELLVGVHATYPVTRKAEGVKMGQGKGAIDHFVARVPAGRVLFHIPQT 116
S L+G + P +G+ MG G G H RV + R LF +P+T
Sbjct 93 STNALIGNYKI-PYIAIIDGITMGGGVGLSVHGKYRVASDRTLFAMPET 140
> CE02372
Length=2207
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 23/86 (26%), Positives = 44/86 (51%), Gaps = 9/86 (10%)
Query 8 PPVR-FKAPQQVRIKCPKGRIVPLNEPEVKLGKTIVAAAPHRVTEAQLEQARRTLRRYLG 66
PP+ F+ + ++CP G + EP+ +L ++ P ++ +E +RT+ Y G
Sbjct 587 PPIDPFREQIVMSLRCPIGPESNMLEPDTELASRLILEQP-VLSMVDMEVIKRTM--YKG 643
Query 67 -RSKELLVGVHATYPVTRKAEGVKMG 91
++KE+ + TYPV +G+ G
Sbjct 644 WKAKEIDI----TYPVKYGVKGLVPG 665
> 7291456
Length=121
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 22/62 (35%), Positives = 31/62 (50%), Gaps = 5/62 (8%)
Query 4 GFKMPPVRFKAPQQVRIKCPKGRIVPLNEPEVKLGKT-IVAAAPH----RVTEAQLEQAR 58
GF+ PP APQQV + +G+ V L P LG+ VA P R T+ + E +
Sbjct 16 GFQAPPSYDSAPQQVYPQLHQGQGVILQAPPNVLGQCPSVATCPSCGVRRETKVEFEPST 75
Query 59 RT 60
+T
Sbjct 76 KT 77
> 7293222
Length=2354
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 12 FKAPQQVRIKCPKGRIVPLNEPEVKLGK 39
FK ++ + CPK R +P P + LGK
Sbjct 425 FKVKDRIELYCPKCRPLPKKLPGIYLGK 452
> At3g09550
Length=436
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query 16 QQVRIKCPKGRIVPLNEPEVKLGKTIVAAAPHRVTEAQLEQARRTLRRYLGRSKEL 71
+++ +C + LN+P +L KT+ + QLEQ R+T + G +KEL
Sbjct 197 KEILSRCGALKANELNQPRDELRKTVTEI--KKDVHTQLEQTRKTNKNVDGIAKEL 250
> At3g32917
Length=757
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query 4 GFKMPPVRFKAPQQVRIKCPKGRIVPLNEPEVKLGKTIVAAAPHRVTEAQLEQARRTLRR 63
G + PV P+ V P R P E++ G ++ AP+R+ A++ + ++ L
Sbjct 518 GLRGLPVTISMPESVGQGLPPSRSDPFT-IELEPGTASLSKAPYRMAPAEMAELKKQLED 576
Query 64 YLGR 67
LG+
Sbjct 577 LLGK 580
Lambda K H
0.322 0.139 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40