bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3311_orf1
Length=196
Score E
Sequences producing significant alignments: (Bits) Value
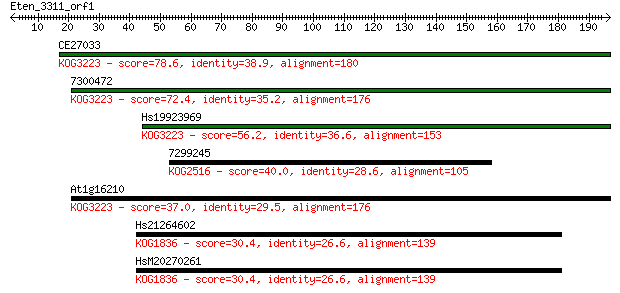
CE27033 78.6 7e-15
7300472 72.4 6e-13
Hs19923969 56.2 4e-08
7299245 40.0 0.003
At1g16210 37.0 0.025
Hs21264602 30.4 2.4
HsM20270261 30.4 2.5
> CE27033
Length=223
Score = 78.6 bits (192), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 70/192 (36%), Positives = 100/192 (52%), Gaps = 27/192 (14%)
Query 17 MPVRV---NQRAVEARERKAAAAAAKAAAAEQEAERKRWEDNDKLVSRKLERKAESQAKA 73
MP + N + AR+RKA A +A + E +W DNDKL +RK++RK + + K
Sbjct 1 MPKKFASENPKVTAARDRKATAKKDEADKKAKATEDAKWVDNDKLNNRKMQRKEDDEKKR 60
Query 74 EEKMQRKMELRKLADEEMQTLAGNSTKKNQSPTKLTRAQI---------LQRQLRAAAME 124
EE ++RK E RKLA+EEM +L GN + K+TRA I + R+L +
Sbjct 61 EEALRRKEENRKLAEEEMSSL-GNKKPAGAATQKVTRAHIHIRKEDEERINREL-EEKRK 118
Query 125 KEASSPSDEGSVLRLEPNINHVMRAESLQAQLEGKNIVSASNLDDALSQLALSEAGGEDR 184
+EA G +L V L+ + EG A N+DDAL L +A +D+
Sbjct 119 QEAQKIEVAGDLL--------VENLNKLEVE-EG----EARNVDDALKVLGEEKALDDDK 165
Query 185 HPERRMKAAYRA 196
HPE+RM+AAY A
Sbjct 166 HPEKRMRAAYLA 177
> 7300472
Length=213
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 62/179 (34%), Positives = 104/179 (58%), Gaps = 16/179 (8%)
Query 21 VNQRAVEARERKAAAAAAKAAAAEQEAERKRWEDNDKLVSRKLERKAESQAKAEEKMQRK 80
+N +AVEARERK A A +EAE + W D+DK +++K +RK E + K E +RK
Sbjct 7 INSKAVEARERKEATKKATQEKKSKEAEDRLWRDDDKNLAKKQQRKDEEERKRAEAAKRK 66
Query 81 MELRKLADEEMQTLAGNSTKKNQSPTKLTRAQIL---QRQLRAAAMEKEASSPSDEGSVL 137
E + L D+EM ++ +T++ Q K+ R IL +++ R EA+ P V+
Sbjct 67 AEAKALLDQEMSSI---NTQRKQPLAKINRQMILEEMEKKQRVIEAINEANKPMAARVVV 123
Query 138 RLEPNINHVMRAESLQAQLEGKNIVSASNLDDALSQLALSEAGGEDRHPERRMKAAYRA 196
+ NH+ E+L + ++ ASN+D+A+ L+++++ ED+HPE+RM+AAY+
Sbjct 124 Q-----NHI--EENLNRSMADTDV--ASNIDEAIVVLSVNDS-EEDKHPEKRMRAAYKT 172
> Hs19923969
Length=223
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 56/158 (35%), Positives = 88/158 (55%), Gaps = 18/158 (11%)
Query 44 EQEAERKRWEDNDKLVSRKLERKAESQAKAEEKMQRKMELRKLADEEMQTLAGNSTKKNQ 103
++E E W+D+DK V RK +RK E + + ++++RK E ++L +EE L G +
Sbjct 31 QKELEDAYWKDDDKHVMRKEQRKEEKEKRRLDQLERKKETQRLLEEEDSKLKGGKAPRVA 90
Query 104 SPTKLTRAQI---LQR--QLRAAAMEKEASSPSDEGSVLRLEPNINHVMRAESLQAQLEG 158
+ +K+TRAQI L+R QLR A E + E + LE N+N + E
Sbjct 91 TSSKVTRAQIEDTLRRDHQLREAPDTAEKAKSHLE---VPLEENVNRRVLEEG------- 140
Query 159 KNIVSASNLDDALSQLALSEAGGEDRHPERRMKAAYRA 196
V A ++DA++ L+++E DRHPERRM+AA+ A
Sbjct 141 --SVEARTIEDAIAVLSVAEEAA-DRHPERRMRAAFTA 175
> 7299245
Length=678
Score = 40.0 bits (92), Expect = 0.003, Method: Composition-based stats.
Identities = 30/114 (26%), Positives = 61/114 (53%), Gaps = 12/114 (10%)
Query 53 EDNDKLVSR------KLERKAESQAKAEEKMQRKMELRKLADEEMQTLA-GNSTKKNQSP 105
+D+D +VS +LE E A AE + L+++ +E++TLA G +TKK+++
Sbjct 530 DDDDGIVSTVEEMSLELETDTEPHAGAETI---ETPLKEINFQELRTLALGQATKKSRAA 586
Query 106 TKLTRAQILQRQLRAAAMEKEASSPSDEGSVLR--LEPNINHVMRAESLQAQLE 157
TK+ QI+++ RA + E S + + + L ++ +++ E ++ +E
Sbjct 587 TKMKIRQIIEQHYRAKGKQIENDSAETQKTQGKAGLRQSVKSIIKQEKIKEMIE 640
> At1g16210
Length=230
Score = 37.0 bits (84), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 52/193 (26%), Positives = 97/193 (50%), Gaps = 29/193 (15%)
Query 21 VNQRAVEARERKAAAAAAKAAAAEQEAERKRWEDNDKLVSRKLERKAESQAKAEEKMQRK 80
+N +A A+ RK AA A + +E E + W + + S+ ++++ E K E +K
Sbjct 3 LNSKAEVAKSRKNAAEAEQKDRQTREKEEQYWREAEGPKSKAVKKREEEAEKKAETAAKK 62
Query 81 MELRKLADEEMQTLAGNSTKKNQS------PT-KLTRAQIL-QRQLRAAAMEKEASSPS- 131
+E ++LA++E + L K ++ P K+T A+++ +R+ A+ K+A
Sbjct 63 LEAKRLAEQEEKELEKALKKPDKKANRVTVPVPKVTEAELIRRREEDQVALAKKAEDSKK 122
Query 132 --------DEGSVLRLEPNINHVMRAESLQAQLEGKNIVSASNLDDALSQLALSEAGGED 183
DE + L N N R +SL + A +D+AL+++ +S+ D
Sbjct 123 KQTRMAGEDEYEKMVLVTNTN---RDDSL---------IEAHTVDEALARITVSDNLPVD 170
Query 184 RHPERRMKAAYRA 196
RHPE+R+KA+++A
Sbjct 171 RHPEKRLKASFKA 183
> Hs21264602
Length=3695
Score = 30.4 bits (67), Expect = 2.4, Method: Composition-based stats.
Identities = 37/144 (25%), Positives = 57/144 (39%), Gaps = 10/144 (6%)
Query 42 AAEQEAERKRWEDNDKLVSRKLERKAESQAKAEEKMQRKMELRKLADEEMQTLAGNSTKK 101
A QE WE+N L ++ +R A+ +A M + L + D + NS +
Sbjct 2350 ARVQEQLSSLWEENQALATQTRDRLAQHEAGL---MDLREALNRAVDATREAQELNSRNQ 2406
Query 102 NQSPTKLTRAQILQRQLRAAAMEKEASSPSDE-GSVLRLEPNINHVMRA-ESLQAQLEGK 159
+ L R Q L R A ++ + D SV RL +++ E L A L+G
Sbjct 2407 ERLEEALQRKQELSRD--NATLQATLHAARDTLASVFRLLHSLDQAKEELERLAASLDGA 2464
Query 160 N---IVSASNLDDALSQLALSEAG 180
+ A S+L L EA
Sbjct 2465 RTPLLQRMQTFSPAGSKLRLVEAA 2488
> HsM20270261
Length=3695
Score = 30.4 bits (67), Expect = 2.5, Method: Composition-based stats.
Identities = 37/144 (25%), Positives = 57/144 (39%), Gaps = 10/144 (6%)
Query 42 AAEQEAERKRWEDNDKLVSRKLERKAESQAKAEEKMQRKMELRKLADEEMQTLAGNSTKK 101
A QE WE+N L ++ +R A+ +A M + L + D + NS +
Sbjct 2350 ARVQEQLSSLWEENQALATQTRDRLAQHEAGL---MDLREALNRAVDATREAQELNSRNQ 2406
Query 102 NQSPTKLTRAQILQRQLRAAAMEKEASSPSDE-GSVLRLEPNINHVMRA-ESLQAQLEGK 159
+ L R Q L R A ++ + D SV RL +++ E L A L+G
Sbjct 2407 ERLEEALQRKQELSRD--NATLQATLHAARDTLASVFRLLHSLDQAKEELERLAASLDGA 2464
Query 160 N---IVSASNLDDALSQLALSEAG 180
+ A S+L L EA
Sbjct 2465 RTPLLQRMQTFSPAGSKLRLVEAA 2488
Lambda K H
0.306 0.117 0.298
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3335884518
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40