bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3347_orf2
Length=114
Score E
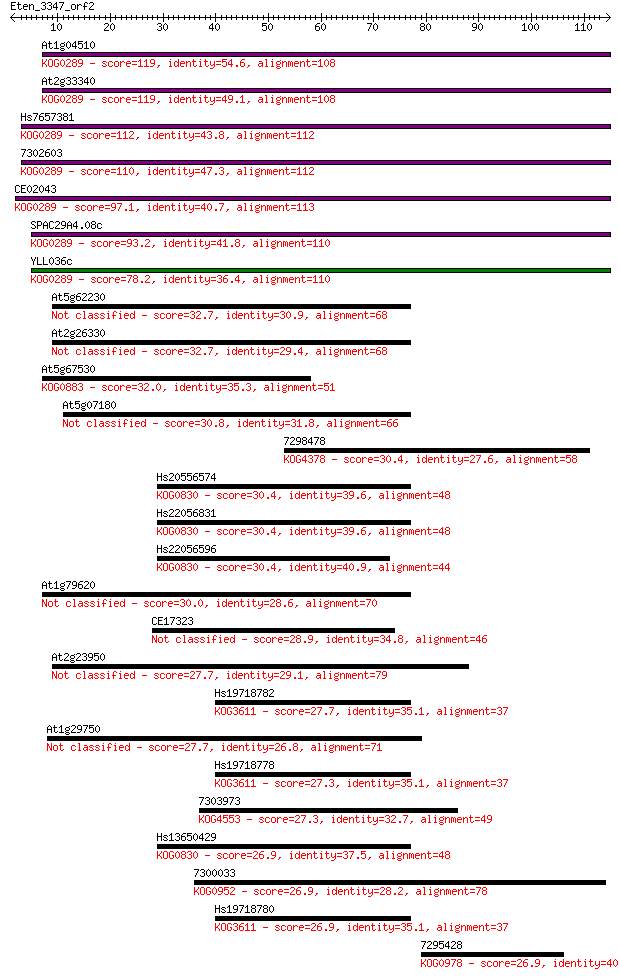
Sequences producing significant alignments: (Bits) Value
At1g04510 119 1e-27
At2g33340 119 2e-27
Hs7657381 112 2e-25
7302603 110 6e-25
CE02043 97.1 8e-21
SPAC29A4.08c 93.2 9e-20
YLL036c 78.2 3e-15
At5g62230 32.7 0.17
At2g26330 32.7 0.18
At5g67530 32.0 0.27
At5g07180 30.8 0.56
7298478 30.4 0.80
Hs20556574 30.4 0.81
Hs22056831 30.4 0.83
Hs22056596 30.4 0.91
At1g79620 30.0 1.1
CE17323 28.9 2.5
At2g23950 27.7 5.1
Hs19718782 27.7 6.1
At1g29750 27.7 6.1
Hs19718778 27.3 6.6
7303973 27.3 7.3
Hs13650429 26.9 8.2
7300033 26.9 8.5
Hs19718780 26.9 9.3
7295428 26.9 9.7
> At1g04510
Length=535
Score = 119 bits (299), Expect = 1e-27, Method: Composition-based stats.
Identities = 59/120 (49%), Positives = 74/120 (61%), Gaps = 12/120 (10%)
Query 7 CAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPR 66
CA++GEVP+E V K+ L+YEKRLI HI+ G CPV+G D++PIKT VKP+
Sbjct 3 CAISGEVPEEPVVSKKSGLLYEKRLIQTHISDYGKCPVTGEPHTLDDIVPIKTGKIVKPK 62
Query 67 PLSASSIPGLLSCLQT------------EWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
PL +SIPGLL QT EWDSLM F L+ L R++L+HALYQ DA
Sbjct 63 PLHTASIPGLLGTFQTDHIELKWMNLIWEWDSLMLSNFALEQQLHTARQELSHALYQHDA 122
> At2g33340
Length=540
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 53/108 (49%), Positives = 72/108 (66%), Gaps = 0/108 (0%)
Query 7 CAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPR 66
CA++GEVP E V K+ L++E+RLI +HI+ G CPV+G L D++PIKT +KP+
Sbjct 3 CAISGEVPVEPVVSTKSGLLFERRLIERHISDYGKCPVTGEPLTIDDIVPIKTGEIIKPK 62
Query 67 PLSASSIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
L +SIPGLL Q EWD LM F L+ L R++L+HALYQ D+
Sbjct 63 TLHTASIPGLLGTFQNEWDGLMLSNFALEQQLHTARQELSHALYQHDS 110
> Hs7657381
Length=504
Score = 112 bits (279), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 49/112 (43%), Positives = 76/112 (67%), Gaps = 0/112 (0%)
Query 3 MSLVCAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPT 62
MSL+C+++ EVP+ P +N +YE+RLI K+IA G P++ L ++ L+ IK
Sbjct 1 MSLICSISNEVPEHPCVSPVSNHVYERRLIEKYIAENGTDPINNQPLSEEQLIDIKVAHP 60
Query 63 VKPRPLSASSIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
++P+P SA+SIP +L LQ EWD++M +F L+ L+ R++L+HALYQ DA
Sbjct 61 IRPKPPSATSIPAILKALQDEWDAVMLHSFTLRQQLQTTRQELSHALYQHDA 112
> 7302603
Length=505
Score = 110 bits (275), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 53/112 (47%), Positives = 78/112 (69%), Gaps = 0/112 (0%)
Query 3 MSLVCAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPT 62
M+LVCA+T EVP+ V P + ++EKR+I K++ GC P+SG +L+ ++L+ IKT
Sbjct 1 MALVCALTNEVPETPVVSPHSGAVFEKRVIEKYLLENGCDPISGKELKPEELIEIKTPAV 60
Query 63 VKPRPLSASSIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
VKP+P SA+SIP L +Q EWD+LM +F + L+ R++L+HALYQ DA
Sbjct 61 VKPKPPSATSIPATLKTMQDEWDALMIHSFTQRQQLQTTRQELSHALYQHDA 112
> CE02043
Length=509
Score = 97.1 bits (240), Expect = 8e-21, Method: Composition-based stats.
Identities = 46/114 (40%), Positives = 74/114 (64%), Gaps = 1/114 (0%)
Query 2 KMSLVCAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNP 61
+MS VC ++GE+ ++ V + I+++RLI+K IA G P+S +L + L+ +K+
Sbjct 17 EMSFVCGISGELTEDPVVSQVSGHIFDRRLIVKFIAENGTDPISHGELSEDQLVSLKSGG 76
Query 62 T-VKPRPLSASSIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
T PR +S +SIP LL LQ EWD++M +F L+ L+ R++L+H+LYQ DA
Sbjct 77 TGSAPRNVSGTSIPSLLKMLQDEWDTVMLNSFSLRQQLQIARQELSHSLYQHDA 130
> SPAC29A4.08c
Length=488
Score = 93.2 bits (230), Expect = 9e-20, Method: Compositional matrix adjust.
Identities = 46/110 (41%), Positives = 69/110 (62%), Gaps = 0/110 (0%)
Query 5 LVCAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVK 64
+ C+++GE P E V + +YEKRLI + I T PV+ + +DL+P+K V+
Sbjct 1 MFCSISGETPKEPVISRVSGNVYEKRLIEQVIRETSKDPVTQQECTLEDLVPVKVPDFVR 60
Query 65 PRPLSASSIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
PRP SA+S+P LLS Q EWDS+ E F L+ +L + +++L+ ALY DA
Sbjct 61 PRPPSATSLPALLSLFQEEWDSVALEQFELRRNLTETKQELSTALYSLDA 110
> YLL036c
Length=503
Score = 78.2 bits (191), Expect = 3e-15, Method: Composition-based stats.
Identities = 40/120 (33%), Positives = 72/120 (60%), Gaps = 12/120 (10%)
Query 5 LVCAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVK 64
++CA++G+VP V PK+ I+EK L+ +++ TG P++ L ++++ I P+ +
Sbjct 1 MLCAISGKVPRRPVLSPKSRTIFEKSLLEQYVKDTGNDPITNEPLSIEEIVEIV--PSAQ 58
Query 65 PRPLSAS----------SIPGLLSCLQTEWDSLMSEAFLLKTHLEQVRKQLAHALYQQDA 114
L+ S SIP LL+ LQ EWD++M E F L++ L+ + K+L+ +Y++DA
Sbjct 59 QASLTESTNSATLKANYSIPNLLTSLQNEWDAIMLENFKLRSTLDSLTKKLSTVMYERDA 118
> At5g62230
Length=966
Score = 32.7 bits (73), Expect = 0.17, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 34/68 (50%), Gaps = 5/68 (7%)
Query 9 VTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPRPL 68
+ G++PDE C +L+Y L + G P S ++L+Q + L +K N P P
Sbjct 107 LAGQIPDEIGNC--ASLVY---LDLSENLLYGDIPFSISKLKQLETLNLKNNQLTGPVPA 161
Query 69 SASSIPGL 76
+ + IP L
Sbjct 162 TLTQIPNL 169
> At2g26330
Length=976
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 20/68 (29%), Positives = 34/68 (50%), Gaps = 5/68 (7%)
Query 9 VTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPRPL 68
++G++PDE C + L + +G P S ++L+Q + L +K N + P P
Sbjct 104 LSGQIPDEIGDCSSL-----QNLDLSFNELSGDIPFSISKLKQLEQLILKNNQLIGPIPS 158
Query 69 SASSIPGL 76
+ S IP L
Sbjct 159 TLSQIPNL 166
> At5g67530
Length=595
Score = 32.0 bits (71), Expect = 0.27, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 7 CAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPI 57
CA+T +P E C ++E I+ +I G PV+GA L+ +DL+P+
Sbjct 42 CALTF-LPFEDPVCTIDGSVFEITTIVPYIRKFGKHPVTGAPLKGEDLIPL 91
> At5g07180
Length=932
Score = 30.8 bits (68), Expect = 0.56, Method: Composition-based stats.
Identities = 21/73 (28%), Positives = 32/73 (43%), Gaps = 19/73 (26%)
Query 11 GEVPDEAVFCPK-------TNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTV 63
G++PDE C TNL++ G P S ++L+Q + L +K N
Sbjct 76 GQIPDEIGNCVSLAYVDFSTNLLF------------GDIPFSISKLKQLEFLNLKNNQLT 123
Query 64 KPRPLSASSIPGL 76
P P + + IP L
Sbjct 124 GPIPATLTQIPNL 136
> 7298478
Length=644
Score = 30.4 bits (67), Expect = 0.80, Method: Compositional matrix adjust.
Identities = 16/59 (27%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 53 DLLPIKTNPTVKPRPLSASSIPGLLSCLQTEWDSLMSE-AFLLKTHLEQVRKQLAHALY 110
++ P +NP +KP+ L A S GL + + +W SE + E+V ++L + +
Sbjct 517 NITPQSSNPLLKPQQLLAKSSSGLSAQMDADWRKEFSELRDFVDQRCEKVERELEYVTF 575
> Hs20556574
Length=288
Score = 30.4 bits (67), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 11/59 (18%)
Query 29 KRLIMKHIAATGCCPVSG--------AQLQ---QQDLLPIKTNPTVKPRPLSASSIPGL 76
+R ++K AATG P++G +Q+Q ++ LP+ T+P V +PL +S L
Sbjct 97 QRAVLKFAAATGATPIAGCFTPGTFTSQIQAAFREPRLPVVTDPRVDHQPLMEASYVNL 155
> Hs22056831
Length=288
Score = 30.4 bits (67), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 32/59 (54%), Gaps = 11/59 (18%)
Query 29 KRLIMKHIAATGCCPVSG--------AQLQ---QQDLLPIKTNPTVKPRPLSASSIPGL 76
+R ++K AATG P++G +Q+Q ++ LP+ T+P V +PL +S L
Sbjct 97 QRAVLKFAAATGATPIAGCFTPGTFTSQIQAAFREPRLPVVTDPRVDHQPLMEASYVNL 155
> Hs22056596
Length=247
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 31/55 (56%), Gaps = 11/55 (20%)
Query 29 KRLIMKHIAATGCCPVSG--------AQLQ---QQDLLPIKTNPTVKPRPLSASS 72
+R ++K AATG P++G +Q+Q ++ LP+ T+P V +PL +S
Sbjct 56 QRAVLKFAAATGATPIAGCFTPGTFTSQIQAAFREPRLPVVTDPRVDHQPLMEAS 110
> At1g79620
Length=980
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 20/70 (28%), Positives = 29/70 (41%), Gaps = 5/70 (7%)
Query 7 CAVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPR 66
C TG +P+E + + L + TG P S L + L + N P
Sbjct 132 CGFTGTIPNELGYLKDLSF-----LALNSNNFTGKIPASLGNLTKVYWLDLADNQLTGPI 186
Query 67 PLSASSIPGL 76
P+S+ S PGL
Sbjct 187 PISSGSSPGL 196
> CE17323
Length=532
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query 28 EKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPRPLSASSI 73
EKR I+K+ + G + GA+ + D L +T V RP++++SI
Sbjct 99 EKRRILKYASKDGKFKIEGARSEFMDFLIQQTRDAV--RPMTSTSI 142
> At2g23950
Length=607
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 6/79 (7%)
Query 9 VTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPRPL 68
++G++P E PK + L + + +G P S QL L + N P P
Sbjct 83 ISGKIPPEICSLPKL-----QTLDLSNNRFSGEIPGSVNQLSNLQYLRLNNNSLSGPFPA 137
Query 69 SASSIPGLLSCLQTEWDSL 87
S S IP LS L +++L
Sbjct 138 SLSQIPH-LSFLDLSYNNL 155
> Hs19718782
Length=682
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 15/37 (40%), Gaps = 0/37 (0%)
Query 40 GCCPVSGAQLQQQDLLPIKTNPTVKPRPLSASSIPGL 76
GCC G Q LP VK PL ++P L
Sbjct 376 GCCAAPGMQYNASSALPDDILNFVKTHPLMDEAVPSL 412
> At1g29750
Length=913
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 19/71 (26%), Positives = 32/71 (45%), Gaps = 5/71 (7%)
Query 8 AVTGEVPDEAVFCPKTNLIYEKRLIMKHIAATGCCPVSGAQLQQQDLLPIKTNPTVKPRP 67
A +G +P E NL++ K+L++ TG P S A+LQ + + P P
Sbjct 134 AFSGTIPQEL-----GNLVHLKKLLLSSNKLTGTLPASLARLQNMTDFEMIASGLTGPIP 188
Query 68 LSASSIPGLLS 78
S + L++
Sbjct 189 SVISVLSNLVN 199
> Hs19718778
Length=517
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 15/37 (40%), Gaps = 0/37 (0%)
Query 40 GCCPVSGAQLQQQDLLPIKTNPTVKPRPLSASSIPGL 76
GCC G Q LP VK PL ++P L
Sbjct 376 GCCAAPGMQYNASSALPDDILNFVKTHPLMDEAVPSL 412
> 7303973
Length=1428
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 26/52 (50%), Gaps = 3/52 (5%)
Query 37 AATGCC---PVSGAQLQQQDLLPIKTNPTVKPRPLSASSIPGLLSCLQTEWD 85
+AT C +SG + Q+ L ++NP L+ I L +CLQ ++D
Sbjct 532 SATSFCTQHSISGYSIMTQNTLGSRSNPQRNFDMLTLEIIGMLRNCLQQQFD 583
> Hs13650429
Length=169
Score = 26.9 bits (58), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 30/59 (50%), Gaps = 11/59 (18%)
Query 29 KRLIMKHIAATGCCPVSG--------AQLQ---QQDLLPIKTNPTVKPRPLSASSIPGL 76
+R ++K AATG P++G Q+Q ++ L + T+P +PL+ SS L
Sbjct 84 QRAVLKFAAATGATPIAGHFTPGTFTNQIQAAFREPRLLVVTDPRADHQPLTESSYVNL 142
> 7300033
Length=1809
Score = 26.9 bits (58), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 36/78 (46%), Gaps = 12/78 (15%)
Query 36 IAATGCCPVSGAQLQQQDLLPIKTNPTVKPRPLSASSIPGLLSCLQTEWDSLMSEAFLLK 95
+ +T C P+S L +LP P + PL P +SCL+ +++ E+
Sbjct 913 LGSTTCIPLSFQHL----VLPEHHPPLTELLPLR----PLPVSCLK----NVVYESLYKF 960
Query 96 THLEQVRKQLAHALYQQD 113
TH ++ Q+ H LY D
Sbjct 961 THFNPIQTQIFHCLYHTD 978
> Hs19718780
Length=888
Score = 26.9 bits (58), Expect = 9.3, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 15/37 (40%), Gaps = 0/37 (0%)
Query 40 GCCPVSGAQLQQQDLLPIKTNPTVKPRPLSASSIPGL 76
GCC G Q LP VK PL ++P L
Sbjct 376 GCCAAPGMQYNASSALPDDILNFVKTHPLMDEAVPSL 412
> 7295428
Length=1031
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 11/27 (40%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 79 CLQTEWDSLMSEAFLLKTHLEQVRKQL 105
CLQ+++ L +E+ +KT L++ R QL
Sbjct 399 CLQSQFSVLYNESMQIKTMLDETRNQL 425
Lambda K H
0.319 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40