bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3361_orf1
Length=258
Score E
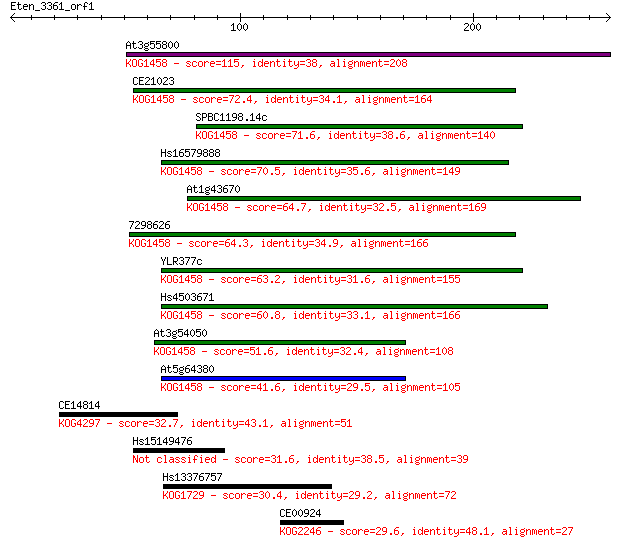
Sequences producing significant alignments: (Bits) Value
At3g55800 115 1e-25
CE21023 72.4 8e-13
SPBC1198.14c 71.6 1e-12
Hs16579888 70.5 3e-12
At1g43670 64.7 2e-10
7298626 64.3 2e-10
YLR377c 63.2 5e-10
Hs4503671 60.8 3e-09
At3g54050 51.6 2e-06
At5g64380 41.6 0.001
CE14814 32.7 0.79
Hs15149476 31.6 1.9
Hs13376757 30.4 4.0
CE00924 29.6 5.9
> At3g55800
Length=393
Score = 115 bits (287), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 79/212 (37%), Positives = 111/212 (52%), Gaps = 17/212 (8%)
Query 51 GMTLQELLDRLSLHTDLRSALPTLFEACGCISKALRSTKVTKSGTSNSFGEEQLSVDLLA 110
G +L+E L + + LR+ L + EA I+ +R+ + NSFG+EQL+VD+LA
Sbjct 77 GQSLEEFLAQATPDKGLRTLLMCMGEALRTIAFKVRTASCGGTACVNSFGDEQLAVDMLA 136
Query 111 ERQLRDWAKRCSVVKAISSEEETALQEVNA--NGSLVVCWDPLDGSSIIDCNWSVASIFG 168
++ L + + V K SEE LQ++ G V +DPLDGSSI+D N++V +IFG
Sbjct 137 DKLLFEALQYSHVCKYACSEEVPELQDMGGPVEGGFSVAFDPLDGSSIVDTNFTVGTIFG 196
Query 169 IWRIGENGLQWNGPDTLINSTGRQQVAAVLVIYGPRTTALLSVCGV--TVDLQLDAEGGA 226
+W D L TG QVAA + IYGPRTT +L+V G T + L EG
Sbjct 197 VWP----------GDKLTGITGGDQVAAAMGIYGPRTTYVLAVKGFPGTHEFLLLDEGKW 246
Query 227 PVILRGPCCIKKNGAKIFSPANLRAAQDLPAY 258
+ + K+FSP NLRA D Y
Sbjct 247 QHVKE---TTEIAEGKMFSPGNLRATFDNSEY 275
> CE21023
Length=341
Score = 72.4 bits (176), Expect = 8e-13, Method: Compositional matrix adjust.
Identities = 56/176 (31%), Positives = 85/176 (48%), Gaps = 17/176 (9%)
Query 54 LQELLDRLSLHTDLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVDLL 109
LQE +L + L + A I+ A + + K +G +N GEE +D+L
Sbjct 22 LQEQRKHADASGELTALLTNMLVAIKAIASATQKAGLAKLYGIAGATNVQGEEVKKLDVL 81
Query 110 AERQLRDWAKRCSVVKAISSEEETALQEVNAN--GSLVVCWDPLDGSSIIDCNWSVASIF 167
+ + + K + SEE L EV G +V +DPLDGSS IDC S+ +IF
Sbjct 82 SNELMINMLKSSYTTCLLVSEENDELIEVEEQRRGKYIVTFDPLDGSSNIDCLVSIGTIF 141
Query 168 GIWRIGENGLQWNGPDTL--INSTGRQQVAAVLVIYGPRTTALLS----VCGVTVD 217
GI++ + +GP T+ + G++ VAA +YG T +LS V G T+D
Sbjct 142 GIYK-----KRGDGPATVDDVLKPGKEMVAAGYALYGSATMVVLSTGDGVNGFTLD 192
> SPBC1198.14c
Length=344
Score = 71.6 bits (174), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 54/144 (37%), Positives = 72/144 (50%), Gaps = 7/144 (4%)
Query 81 ISKALRSTKVTKSGTSNSFGEEQLSVDLLAERQLRDWAKRCSVVKAISSEEETALQEVNA 140
I KA + SG NS G+EQ +D + K K I SEEE L V++
Sbjct 62 IRKAELVNLIGLSGIVNSTGDEQKKLDKICNDIFITAMKSNGCCKLIVSEEEEDLIVVDS 121
Query 141 NGSLVVCWDPLDGSSIIDCNWSVASIFGIWRIGENGLQWNGPDTLINSTGRQQVAAVLVI 200
NGS V DP+DGSS ID SV +IFGI+++ G Q + D L G++ VAA +
Sbjct 122 NGSYAVTCDPIDGSSNIDAGVSVGTIFGIYKL-RPGSQGDISDVL--RPGKEMVAAGYTM 178
Query 201 YGPRTTALLS----VCGVTVDLQL 220
YG LL+ V G T+D +
Sbjct 179 YGASAHLLLTTGHRVNGFTLDTDI 202
> Hs16579888
Length=338
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 53/156 (33%), Positives = 81/156 (51%), Gaps = 10/156 (6%)
Query 66 DLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVDLLAERQLRDWAKRC 121
+L L +L A IS A+R + +G++N G++ +D+L+ + + K
Sbjct 30 ELTQLLNSLCTAVKAISSAVRKAGIAHLYGIAGSTNVTGDQVKKLDVLSNDLVMNMLKSS 89
Query 122 SVVKAISSEEE--TALQEVNANGSLVVCWDPLDGSSIIDCNWSVASIFGIWRIGENGLQW 179
+ SEE+ + E G VVC+DPLDGSS IDC SV +IFGI+R ++ +
Sbjct 90 FATCVLVSEEDKHAIIVEPEKRGKYVVCFDPLDGSSNIDCLVSVGTIFGIYR-KKSTDEP 148
Query 180 NGPDTLINSTGRQQVAAVLVIYGPRTTALLSV-CGV 214
+ D L GR VAA +YG T +L++ CGV
Sbjct 149 SEKDAL--QPGRNLVAAGYALYGSATMLVLAMDCGV 182
> At1g43670
Length=341
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 55/177 (31%), Positives = 82/177 (46%), Gaps = 18/177 (10%)
Query 77 ACGCISKALRSTKVTKSGTSNSFGEEQLSVDLLAERQLRDW---AKRCSVVKAISSEEET 133
C ++KA + + +G +N GEEQ +D+L+ + + R SV+ + +EE
Sbjct 47 VCSAVNKAGLAKLIGLAGETNIQGEEQKKLDVLSNDVFVNALVSSGRTSVLVS-EEDEEA 105
Query 134 ALQEVNANGSLVVCWDPLDGSSIIDCNWSVASIFGIWRIGENGLQWNGPDTL-INSTGRQ 192
E + G V +DPLDGSS IDC S+ +IFGI+ + + P T + G +
Sbjct 106 TFVEPSKRGKYCVVFDPLDGSSNIDCGVSIGTIFGIYTLD----HTDEPTTADVLKPGNE 161
Query 193 QVAAVLVIYGPRTTALLS----VCGVTVDLQLDAEGGAPVILRGPCCIKKNGAKIFS 245
VAA +YG +LS V G T+D L IL P N I+S
Sbjct 162 MVAAGYCMYGSSCMLVLSTGTGVHGFTLDPSL-----GEFILTHPDIKIPNKGNIYS 213
> 7298626
Length=334
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 58/180 (32%), Positives = 80/180 (44%), Gaps = 21/180 (11%)
Query 52 MTLQELLDRLSLHTDLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVD 107
LQE S DL L ++ A S A+R + K +G N GEE +D
Sbjct 18 FVLQEQRKFKSATGDLSQLLNSIQTAIKATSSAVRKAGIAKLHGFAGDVNVQGEEVKKLD 77
Query 108 LLAER----QLRDWAKRCSVVKAISSEEETALQEVNANGSLVVCWDPLDGSSIIDCNWSV 163
+L+ L+ C +V + E G +VC+DPLDGSS IDC S+
Sbjct 78 VLSNELFINMLKSSYTTCLMVSEENENVIEVEVE--KQGKYIVCFDPLDGSSNIDCLVSI 135
Query 164 ASIFGIWRIGENGLQWNGPDTLINS--TGRQQVAAVLVIYGPRTTALL----SVCGVTVD 217
SIF I+R + +GP T+ ++ G Q VAA +YG T +L V G T D
Sbjct 136 GSIFAIYR-----KKSDGPPTVEDALQPGNQLVAAGYALYGSATAIVLGLGSGVNGFTYD 190
> YLR377c
Length=348
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 49/164 (29%), Positives = 79/164 (48%), Gaps = 12/164 (7%)
Query 66 DLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVDLLAERQLRDWAKRC 121
D L L A +S +R ++ +G SN G++Q +D+L + + +
Sbjct 40 DFTLVLNALQFAFKFVSHTIRRAELVNLVGLAGASNFTGDQQKKLDVLGDEIFINAMRAS 99
Query 122 SVVKAISSEEETALQEVNAN-GSLVVCWDPLDGSSIIDCNWSVASIFGIWRIGENGLQWN 180
++K + SEE+ L N GS VC DP+DGSS +D SV +I I+R+ + +
Sbjct 100 GIIKVLVSEEQEDLIVFPTNTGSYAVCCDPIDGSSNLDAGVSVGTIASIFRLLPDS---S 156
Query 181 GPDTLINSTGRQQVAAVLVIYGPRTTALLS----VCGVTVDLQL 220
G + G++ VAA +YG T +L+ V G T+D L
Sbjct 157 GTINDVLRCGKEMVAACYAMYGSSTHLVLTLGDGVDGFTLDTNL 200
> Hs4503671
Length=339
Score = 60.8 bits (146), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 55/173 (31%), Positives = 81/173 (46%), Gaps = 11/173 (6%)
Query 66 DLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVDLLAERQLRDWAKRC 121
+L L ++ A IS A+R + +G+ N G+E +D+L+ + + +
Sbjct 30 ELTQLLNSMLTAIKAISSAVRKAGLAHLYGIAGSVNVTGDEVKKLDVLSNSLVINMLQSS 89
Query 122 SVVKAISSEE--ETALQEVNANGSLVVCWDPLDGSSIIDCNWSVASIFGIWRIGENGLQW 179
+ SEE + + G VVC+DPLDGSS IDC S+ +IF I+R +
Sbjct 90 YSTCVLVSEENKDAIITAKEKRGKYVVCFDPLDGSSNIDCLASIGTIFAIYRKTSED-EP 148
Query 180 NGPDTLINSTGRQQVAAVLVIYGPRTTALLSVCGVTVDL-QLDAEGGAPVILR 231
+ D L GR VAA +YG T LS G VDL LD G V++
Sbjct 149 SEKDAL--QCGRNIVAAGYALYGSATLVALST-GQGVDLFMLDPALGEFVLVE 198
> At3g54050
Length=417
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 35/115 (30%), Positives = 60/115 (52%), Gaps = 8/115 (6%)
Query 63 LHTDLRSALPTLFEACGCISKALRSTKVTK----SGTSNSFGEEQLSVDLLAERQLRDWA 118
+ +L + ++ AC I+ ++ ++ G N GE+Q +D+++ +
Sbjct 96 IDAELTIVMSSISLACKQIASLVQRAGISNLTGVQGAVNIQGEDQKKLDVISNEVFSNCL 155
Query 119 KRCSVVKAISSEEE---TALQEVNANGSLVVCWDPLDGSSIIDCNWSVASIFGIW 170
+ I+SEEE A++E + +G+ VV +DPLDGSS ID S SIFGI+
Sbjct 156 RSSGRTGIIASEEEDVPVAVEE-SYSGNYVVVFDPLDGSSNIDAAVSTGSIFGIY 209
> At5g64380
Length=404
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 52/111 (46%), Gaps = 6/111 (5%)
Query 66 DLRSALPTLFEACGCISKALRST------KVTKSGTSNSFGEEQLSVDLLAERQLRDWAK 119
DL L L AC I+ + S K++ + +S S + +D+++ + +
Sbjct 89 DLVVLLYHLQHACKRIASLVASPFNSSLGKLSVNSSSGSDRDAPKPLDIVSNDIVLSSLR 148
Query 120 RCSVVKAISSEEETALQEVNANGSLVVCWDPLDGSSIIDCNWSVASIFGIW 170
V ++SEE + + +G VV DPLDGS ID + +IFGI+
Sbjct 149 NSGKVAVMASEENDSPTWIKDDGPYVVVVDPLDGSRNIDASIPTGTIFGIY 199
> CE14814
Length=351
Score = 32.7 bits (73), Expect = 0.79, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 31/58 (53%), Gaps = 7/58 (12%)
Query 22 WAYF---AGGYLIQSFSSNHNAKMTGKEADFA-GMT---LQELLDRLSLHTDLRSALP 72
W +F +GG+ I+ F+ NHNAK+ + A A G T LQ D L + L S +P
Sbjct 63 WTFFNRPSGGWCIRVFTGNHNAKVDAEGACAAVGATLTGLQNKADALFIQNALLSQIP 120
> Hs15149476
Length=660
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 15/45 (33%), Positives = 26/45 (57%), Gaps = 6/45 (13%)
Query 54 LQELLDRLSLHT------DLRSALPTLFEACGCISKALRSTKVTK 92
LQ++LD L LHT +L +A +++C C+ K ++ K+ K
Sbjct 585 LQKILDDLFLHTLCDYIYELATAFTEFYDSCYCVEKDRQTGKILK 629
> Hs13376757
Length=310
Score = 30.4 bits (67), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 21/81 (25%), Positives = 36/81 (44%), Gaps = 9/81 (11%)
Query 67 LRSALPTLFEACGCISKALRSTKVTKSGTSNSFGEEQLSVDLLA---------ERQLRDW 117
L+ L L E C + K L+S K + E+ + LL +++LR+
Sbjct 135 LQLQLSQLHEQCSSLEKELKSEKEQRQALQRELQHEKDTSSLLRMELQQVEGLKKELREL 194
Query 118 AKRCSVVKAISSEEETALQEV 138
+ ++ I E+E ALQE+
Sbjct 195 QDEKAELQKICEEQEQALQEM 215
> CE00924
Length=419
Score = 29.6 bits (65), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 17/27 (62%), Gaps = 0/27 (0%)
Query 117 WAKRCSVVKAISSEEETALQEVNANGS 143
WAKRC+ +SSEE+ L +N N S
Sbjct 158 WAKRCNKYVFMSSEEDAELPAINLNVS 184
Lambda K H
0.318 0.132 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5400706832
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40