bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3366_orf1
Length=169
Score E
Sequences producing significant alignments: (Bits) Value
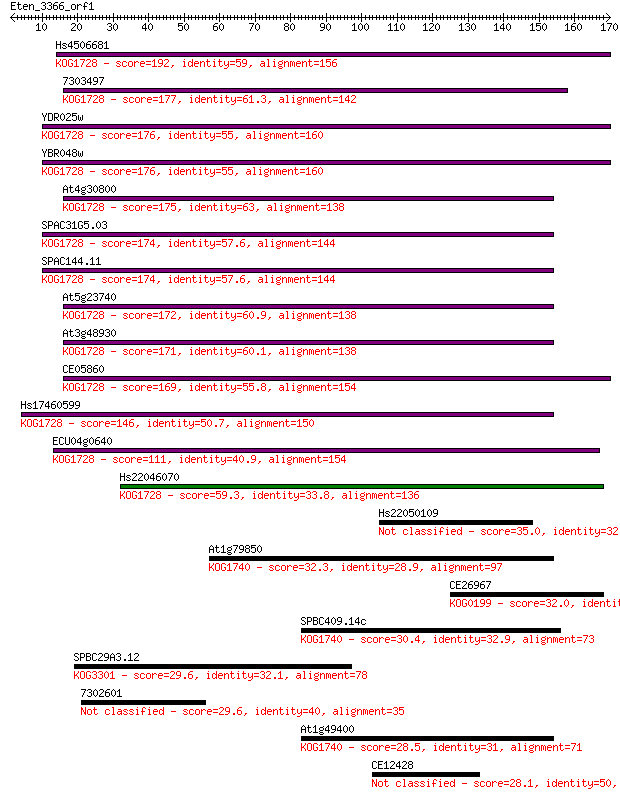
Hs4506681 192 2e-49
7303497 177 1e-44
YDR025w 176 2e-44
YBR048w 176 2e-44
At4g30800 175 4e-44
SPAC31G5.03 174 1e-43
SPAC144.11 174 1e-43
At5g23740 172 2e-43
At3g48930 171 8e-43
CE05860 169 3e-42
Hs17460599 146 2e-35
ECU04g0640 111 6e-25
Hs22046070 59.3 4e-09
Hs22050109 35.0 0.071
At1g79850 32.3 0.40
CE26967 32.0 0.60
SPBC409.14c 30.4 2.0
SPBC29A3.12 29.6 2.9
7302601 29.6 2.9
At1g49400 28.5 7.1
CE12428 28.1 9.0
> Hs4506681
Length=158
Score = 192 bits (489), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 92/160 (57%), Positives = 120/160 (75%), Gaps = 8/160 (5%)
Query 14 DVQTEKAFQRQ----HGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCP 69
D+QTE+A+Q+Q ++ L ++ K++ RY++ +GLGF TP+ A EGTYIDKKCP
Sbjct 3 DIQTERAYQKQPTIFQNKKRVLLGETGKEK-LPRYYKNIGLGFKTPKEAIEGTYIDKKCP 61
Query 70 FTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 129
FTGNVSIRGRI+ G+V KM+R +VIRR YLH++RKY+R EKRHKN++ HLSPCF V+
Sbjct 62 FTGNVSIRGRILSGVVTKMKMQRTIVIRRDYLHYIRKYNRFEKRHKNMSVHLSPCFRDVQ 121
Query 130 EGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 169
GDIVT G+CRPLSKT+RFNVLKV +KQF+ F
Sbjct 122 IGDIVTVGECRPLSKTVRFNVLKVTKA---AGTKKQFQKF 158
> 7303497
Length=155
Score = 177 bits (448), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 87/143 (60%), Positives = 106/143 (74%), Gaps = 1/143 (0%)
Query 16 QTEKAFQRQHGAS-QLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTGNV 74
Q E+AFQ+Q G + + K+ R R+VGLGF TPR A +GTYIDKKCP+TG+V
Sbjct 4 QNERAFQKQFGVNLNRKVKPGITKKKLLRRSRDVGLGFKTPREAIDGTYIDKKCPWTGDV 63
Query 75 SIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIV 134
IRGRI+ G+V AKM+R +VIRR YLHFVRKYSR EKRH+N++ H SP F V+ GDIV
Sbjct 64 RIRGRILTGVVRKAKMQRTIVIRRDYLHFVRKYSRFEKRHRNMSVHCSPVFRDVEHGDIV 123
Query 135 TAGQCRPLSKTIRFNVLKVESNQ 157
T G+CRPLSKT+RFNVLKV Q
Sbjct 124 TIGECRPLSKTVRFNVLKVSKGQ 146
> YDR025w
Length=156
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 88/160 (55%), Positives = 115/160 (71%), Gaps = 5/160 (3%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCP 69
+T+ VQ+E+AFQ+Q ++SK+ R+++ GLGF TP+ A EG+YIDKKCP
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKR--TKRWYKNAGLGFKTPKTAIEGSYIDKKCP 59
Query 70 FTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 129
FTG VSIRG+I+ G V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+
Sbjct 60 FTGLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQ 118
Query 130 EGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 169
GDIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 119 VGDIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> YBR048w
Length=156
Score = 176 bits (446), Expect = 2e-44, Method: Compositional matrix adjust.
Identities = 88/160 (55%), Positives = 115/160 (71%), Gaps = 5/160 (3%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCP 69
+T+ VQ+E+AFQ+Q ++SK+ R+++ GLGF TP+ A EG+YIDKKCP
Sbjct 2 STELTVQSERAFQKQPHIFNNPKVKTSKR--TKRWYKNAGLGFKTPKTAIEGSYIDKKCP 59
Query 70 FTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 129
FTG VSIRG+I+ G V+S KM R +VIRR YLH++ KY+R EKRHKNV H+SP F +V+
Sbjct 60 FTGLVSIRGKILTGTVVSTKMHRTIVIRRAYLHYIPKYNRYEKRHKNVPVHVSPAF-RVQ 118
Query 130 EGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 169
GDIVT GQCRP+SKT+RFNV+KV + G KQF F
Sbjct 119 VGDIVTVGQCRPISKTVRFNVVKVSAAA--GKANKQFAKF 156
> At4g30800
Length=159
Score = 175 bits (443), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 87/141 (61%), Positives = 106/141 (75%), Gaps = 4/141 (2%)
Query 16 QTEKAFQRQHG---ASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTG 72
QTEKAF +Q +S+ + +G R+W+ +GLGF TPR A EGTYID+KCPFTG
Sbjct 4 QTEKAFLKQPKVFLSSKKSGKGKRPGKGGNRFWKNIGLGFKTPREAIEGTYIDQKCPFTG 63
Query 73 NVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 132
VSIRGRI+ G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD
Sbjct 64 TVSIRGRILSGTCHSAKMQRTIIVRRDYLHFVKKYRRYEKRHSNIPAHVSPCF-RVKEGD 122
Query 133 IVTAGQCRPLSKTIRFNVLKV 153
VT GQCRPLSKT+RFNVLKV
Sbjct 123 RVTIGQCRPLSKTVRFNVLKV 143
> SPAC31G5.03
Length=152
Score = 174 bits (440), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 83/144 (57%), Positives = 108/144 (75%), Gaps = 6/144 (4%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCP 69
AT+ VQ+E+AFQ+Q + Q++KK R++++VGLGF TP A G Y+DKKCP
Sbjct 2 ATELVVQSERAFQKQ-----PHIFQNAKKGAGRRWYKDVGLGFKTPAEAIYGEYVDKKCP 56
Query 70 FTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 129
F G VSIRGRI+ G V+S KM R ++IRR YLHF+ KY+R EKRHKN+ H+SP F ++
Sbjct 57 FVGQVSIRGRILTGTVVSTKMHRTIIIRREYLHFIPKYNRYEKRHKNLAAHVSPAF-RIN 115
Query 130 EGDIVTAGQCRPLSKTIRFNVLKV 153
EGD+VT GQCRPLSKT+RFNVL+V
Sbjct 116 EGDVVTVGQCRPLSKTVRFNVLRV 139
> SPAC144.11
Length=152
Score = 174 bits (440), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 83/144 (57%), Positives = 108/144 (75%), Gaps = 6/144 (4%)
Query 10 ATQQDVQTEKAFQRQHGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCP 69
AT+ VQ+E+AFQ+Q + Q++KK R++++VGLGF TP A G Y+DKKCP
Sbjct 2 ATELVVQSERAFQKQ-----PHIFQNAKKGAGRRWYKDVGLGFKTPAEAIYGEYVDKKCP 56
Query 70 FTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVK 129
F G VSIRGRI+ G V+S KM R ++IRR YLHF+ KY+R EKRHKN+ H+SP F ++
Sbjct 57 FVGQVSIRGRILTGTVVSTKMHRTIIIRREYLHFIPKYNRYEKRHKNLAAHVSPAF-RIN 115
Query 130 EGDIVTAGQCRPLSKTIRFNVLKV 153
EGD+VT GQCRPLSKT+RFNVL+V
Sbjct 116 EGDVVTVGQCRPLSKTVRFNVLRV 139
> At5g23740
Length=159
Score = 172 bits (437), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 84/141 (59%), Positives = 104/141 (73%), Gaps = 4/141 (2%)
Query 16 QTEKAFQRQHG---ASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTG 72
QTEKAF +Q +S+++ +G R+W+ +GLGF TPR A +G YID KCPFTG
Sbjct 4 QTEKAFLKQPKVFLSSKISGKGKRPGKGGNRFWKNIGLGFKTPREAIDGAYIDSKCPFTG 63
Query 73 NVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 132
VSIRGRI+ G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD
Sbjct 64 TVSIRGRILAGTCHSAKMQRTIIVRRNYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGD 122
Query 133 IVTAGQCRPLSKTIRFNVLKV 153
V GQCRPLSKT+RFNVLKV
Sbjct 123 HVIIGQCRPLSKTVRFNVLKV 143
> At3g48930
Length=160
Score = 171 bits (432), Expect = 8e-43, Method: Compositional matrix adjust.
Identities = 83/141 (58%), Positives = 104/141 (73%), Gaps = 4/141 (2%)
Query 16 QTEKAFQRQHG---ASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTG 72
QTEKAF +Q +S+ + +G R+W+ +GLGF TPR A +G Y+DKKCPFTG
Sbjct 4 QTEKAFLKQPKVFLSSKKSGKGKRPGKGGNRFWKNIGLGFKTPREAIDGAYVDKKCPFTG 63
Query 73 NVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 132
VSIRGRI+ G SAKM+R +++RR YLHFV+KY R EKRH N+ H+SPCF +VKEGD
Sbjct 64 TVSIRGRILAGTCHSAKMQRTIIVRRDYLHFVKKYQRYEKRHSNIPAHVSPCF-RVKEGD 122
Query 133 IVTAGQCRPLSKTIRFNVLKV 153
+ GQCRPLSKT+RFNVLKV
Sbjct 123 HIIIGQCRPLSKTVRFNVLKV 143
> CE05860
Length=155
Score = 169 bits (428), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 86/157 (54%), Positives = 106/157 (67%), Gaps = 8/157 (5%)
Query 16 QTEKAFQRQHGAS---QLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTG 72
QTE+AF +Q + + + SKK RY REVGLGF PR A EGTYIDKKCP+ G
Sbjct 4 QTERAFLKQPTVNLNNKARILAGSKK--TPRYIREVGLGFKAPRDAVEGTYIDKKCPWAG 61
Query 73 NVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 132
NV IRG I+ G+V+ KM R +V+RR YLH+++KY R EKRHKNV H SP F + GD
Sbjct 62 NVPIRGMILTGVVLKNKMTRTIVVRRDYLHYIKKYRRYEKRHKNVPAHCSPAFRDIHPGD 121
Query 133 IVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFRLF 169
+VT G+CRPLSKT+RFNVLKV + G +K F F
Sbjct 122 LVTIGECRPLSKTVRFNVLKVNKS---GTSKKGFSKF 155
> Hs17460599
Length=329
Score = 146 bits (369), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 76/157 (48%), Positives = 97/157 (61%), Gaps = 16/157 (10%)
Query 4 VQQTMSATQQDVQTEKAFQRQHGASQLALAQSSKKRGKARYWRE-------VGLGFATPR 56
V +AT DV AF + + + ++ R W+E + LGF TP
Sbjct 157 VNWKQAATGPDVGHASAF--------MMITANDEEAPAGRNWQEAPAVLQNINLGFKTPE 208
Query 57 LAKEGTYIDKKCPFTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKN 116
A EGTYIDKKCPFTGN SIRGRI+ GMV KM+R VI R YLH+ KY+ +EK HKN
Sbjct 209 EAIEGTYIDKKCPFTGNASIRGRILFGMVTKMKMQR-TVICRDYLHYTCKYNHLEKHHKN 267
Query 117 VTCHLSPCFEQVKEGDIVTAGQCRPLSKTIRFNVLKV 153
++ +LSPCF ++ DI+T G+C PLSK +RFNVLKV
Sbjct 268 MSVNLSPCFRDIQISDIITVGECWPLSKMVRFNVLKV 304
> ECU04g0640
Length=156
Score = 111 bits (278), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 63/155 (40%), Positives = 90/155 (58%), Gaps = 11/155 (7%)
Query 13 QDVQTEKAFQRQHGASQLALAQSSKKRGKARYWREVGLGFATPRLAKEGTYIDKKCPFTG 72
++V+ K F RQ G A S +++ K R EV GF P A YIDKKCPFTG
Sbjct 2 EEVRKLKVFPRQTGVCAKPYA-SHEEQPKRRVNAEV-CGFKAPETAYTRQYIDKKCPFTG 59
Query 73 NVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFE-QVKEG 131
+V++RGR+ KG+V K ++ +V+ Y+H+ +KY R +RH N + H+SPCFE + G
Sbjct 60 DVNVRGRLFKGVVKKMKAEKTIVVVANYVHYSKKYKRYSRRHTNFSVHMSPCFEGMINVG 119
Query 132 DIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQF 166
D V G+ RPLSKT +S+ V G ++K
Sbjct 120 DTVICGETRPLSKT--------KSSVVLGYIKKAL 146
> Hs22046070
Length=105
Score = 59.3 bits (142), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 46/146 (31%), Positives = 58/146 (39%), Gaps = 63/146 (43%)
Query 32 LAQSSKKRGKARYWREVGLGFATPRL----------AKEGTYIDKKCPFTGNVSIRGRII 81
L SK+ G+ W L P + A EG +IDKKCPFTGNVSIRG
Sbjct 11 LPVESKEAGRGSCWET--LAGEVPVVLQEHQSGLQEAIEGAHIDKKCPFTGNVSIRG--- 65
Query 82 KGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRP 141
V+ DIVT G+C+P
Sbjct 66 ---------------------------------------------DVQISDIVTVGECQP 80
Query 142 LSKTIRFNVLKVESNQVFGNVRKQFR 167
LSKT+RF+VLKV + +KQF+
Sbjct 81 LSKTVRFDVLKVTKA---ASTKKQFQ 103
> Hs22050109
Length=1575
Score = 35.0 bits (79), Expect = 0.071, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 105 RKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPLSKTIR 147
+K SR +++ + CHL PC Q KE ++ CR + +R
Sbjct 1285 KKVSRCFRKYTELFCHLDPCLLQSKESQLLQEENCRKKLEALR 1327
> At1g79850
Length=149
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 28/97 (28%), Positives = 43/97 (44%), Gaps = 2/97 (2%)
Query 57 LAKEGTYIDKKCPFTGNVSIRGRIIKGMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKN 116
L+K ++ + + P V + ++G V+ A + V + L KY R + K
Sbjct 29 LSKPNSFPNHRMPALVPVIRAMKTMQGRVVCATSDKTVAVEVVRLAPHPKYKRRVRMKKK 88
Query 117 VTCHLSPCFEQVKEGDIVTAGQCRPLSKTIRFNVLKV 153
H P Q K GD+V + RP+SKT F L V
Sbjct 89 YQAH-DPD-NQFKVGDVVRLEKSRPISKTKSFVALPV 123
> CE26967
Length=1043
Score = 32.0 bits (71), Expect = 0.60, Method: Composition-based stats.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 125 FEQVKEGDIVTAGQCRPLSKTIRFNVLKVESNQVFGNVRKQFR 167
+ V+E D+++ G RP K +R K+ + +FG V+K F+
Sbjct 42 LQYVEEVDLLSVGMSRPEQKRLRKEYTKMFPSGIFGKVKKAFK 84
> SPBC409.14c
Length=90
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 15/79 (18%)
Query 83 GMVISAKMKRCVVIRRTYLHF---VRKYSRVEKRH---KNVTCHLSPCFEQVKEGDIVTA 136
G+VIS M + +R F V+KY + + ++ C++ GD VT
Sbjct 7 GIVISRAMPKTAKVRVAKEKFHPVVKKYVTQYQNYMVQDDLNCNV---------GDAVTI 57
Query 137 GQCRPLSKTIRFNVLKVES 155
CRP S T RF ++K+ S
Sbjct 58 QPCRPRSATKRFEIIKILS 76
> SPBC29A3.12
Length=192
Score = 29.6 bits (65), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 5/82 (6%)
Query 19 KAFQRQHGASQLALAQSSKKRGKARYWREVGLGFA-TPRLAKEGTYIDKKCP---FTGNV 74
+ F+ ++L LA R K WR V L + R A+E +D+K P F GN
Sbjct 17 RPFESARLDAELKLAGEYGLRNKHEIWR-VALTLSKIRRAARELLTLDEKDPKRLFEGNA 75
Query 75 SIRGRIIKGMVISAKMKRCVVI 96
IR + G++ ++MK V+
Sbjct 76 IIRRLVRLGILDESRMKLDYVL 97
> 7302601
Length=540
Score = 29.6 bits (65), Expect = 2.9, Method: Composition-based stats.
Identities = 14/42 (33%), Positives = 22/42 (52%), Gaps = 7/42 (16%)
Query 21 FQRQHGASQLALAQSSKKRG-------KARYWREVGLGFATP 55
+ ++GA L+ + K+RG + RYWRE G+ F P
Sbjct 202 YDHRNGAFDWDLSMTLKERGGQQICSQEYRYWRETGVAFVFP 243
> At1g49400
Length=116
Score = 28.5 bits (62), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 32/71 (45%), Gaps = 2/71 (2%)
Query 83 GMVISAKMKRCVVIRRTYLHFVRKYSRVEKRHKNVTCHLSPCFEQVKEGDIVTAGQCRPL 142
G V+S KM++ VV+ L + Y+R KR H + GD V RPL
Sbjct 6 GTVVSNKMQKSVVVAVDRLFHNKIYNRYVKRTSKFMAHDDK--DACNIGDRVKLDPSRPL 63
Query 143 SKTIRFNVLKV 153
SK + V ++
Sbjct 64 SKNKHWIVAEI 74
> CE12428
Length=561
Score = 28.1 bits (61), Expect = 9.0, Method: Composition-based stats.
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query 103 FVRKYSRVEKRHKNVTCHLSPCFEQVKEGD 132
FV Y KRH N TCH P F+ +KE D
Sbjct 432 FVTVYKGNMKRHLN-TCHPQPEFKSLKEWD 460
Lambda K H
0.324 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2454498488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40