bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
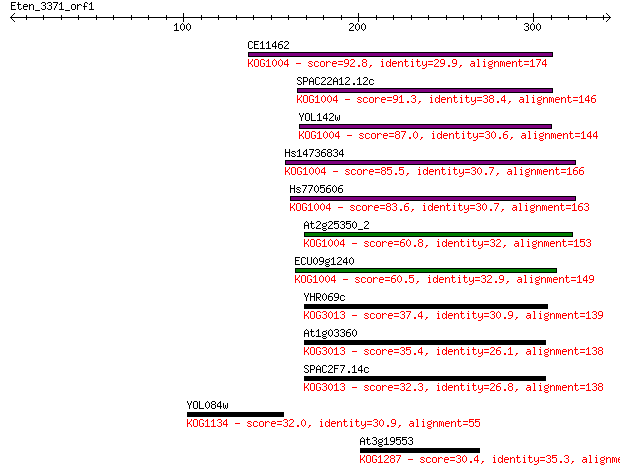
Query= Eten_3371_orf1
Length=343
Score E
Sequences producing significant alignments: (Bits) Value
CE11462 92.8 9e-19
SPAC22A12.12c 91.3 3e-18
YOL142w 87.0 6e-17
Hs14736834 85.5 1e-16
Hs7705606 83.6 6e-16
At2g25350_2 60.8 5e-09
ECU09g1240 60.5 6e-09
YHR069c 37.4 0.041
At1g03360 35.4 0.18
SPAC2F7.14c 32.3 1.7
YOL084w 32.0 1.9
At3g19553 30.4 6.3
> CE11462
Length=226
Score = 92.8 bits (229), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 52/176 (29%), Positives = 89/176 (50%), Gaps = 12/176 (6%)
Query 137 GPCVKARTASMMLPKHVLPPHGPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRS 196
G V+ +T P G ++ + + RY P+ GD V+ I++SK +++ ++I
Sbjct 24 GISVRGQTRIATQPGAFHNDDGKVWLNVHSK-RYIPQEGDRVIAIVTSKTGDFFRLDI-- 80
Query 197 VGPARLATV--AGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSN 254
G A A + FEGAT+R++P L G ++ V +P A++TC
Sbjct 81 -GTAEYAMINFTNFEGATKRNRPNLKTGDIIYATVFDTTPRTEAELTCVDDE------KR 133
Query 255 EKLLGELRGGVIVEVAIPFALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAET 310
+ +G+L GG + +V++ L+ +LQ +G FEI VG+NGR+W+ A T
Sbjct 134 ARGMGQLNGGFMFKVSLNHCRRLINPSCKILQTVGKFFKFEITVGMNGRIWISAST 189
> SPAC22A12.12c
Length=240
Score = 91.3 bits (225), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 56/147 (38%), Positives = 84/147 (57%), Gaps = 7/147 (4%)
Query 165 STNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSV 224
S RY P D V+G I S+ +E Y V+I S A+L +A FE TR+S+P L+VGS+
Sbjct 67 SRTKRYIPATNDQVIGQIISRFAEGYRVDIGSAHIAQLNALA-FENVTRKSRPNLNVGSL 125
Query 225 LLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGG-VIVEVAIPFALTLLCHDSL 283
+ RVS + ++ C ++ K GEL+ G +I +++ L+ +
Sbjct 126 VYARVSLADRDMEPELECFDATTGKAAG-----YGELKNGYMITGLSLSHCRKLILPKNT 180
Query 284 VLQLLGSRLPFEIAVGLNGRVWLRAET 310
+LQ LGS +PFEIAVG+NGRVW+ +E
Sbjct 181 LLQTLGSYIPFEIAVGMNGRVWVNSEN 207
> YOL142w
Length=240
Score = 87.0 bits (214), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 81/145 (55%), Gaps = 6/145 (4%)
Query 166 TNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVL 225
++ RY P V D V+G+I S+ Y V++++ + + F A+++++P L VG ++
Sbjct 60 SSKRYIPSVNDFVIGVIIGTFSDSYKVSLQNFSSSVSLSYMAFPNASKKNRPTLQVGDLV 119
Query 226 LVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSL-V 284
RV L A++ C S+ T + G L G+I++V FA LL ++ +
Sbjct 120 YARVCTAEKELEAEIECFDST-----TGRDAGFGILEDGMIIDVNFNFARQLLFNNDFPL 174
Query 285 LQLLGSRLPFEIAVGLNGRVWLRAE 309
L++L + FE+A+GLNG++W++ E
Sbjct 175 LKVLAAHTKFEVAIGLNGKIWVKCE 199
> Hs14736834
Length=275
Score = 85.5 bits (210), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 51/170 (30%), Positives = 92/170 (54%), Gaps = 14/170 (8%)
Query 158 GPTYVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKP 217
G Y S RY P GD V+GI+++K + + V++ PA L+ ++ FEGAT+R++P
Sbjct 97 GGVYWVDSQQKRYVPVKGDHVIGIVTAKSGDIFKVDVGGSEPASLSYLS-FEGATKRNRP 155
Query 218 QLSVGSVL----LVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPF 273
+ VG ++ +V + P + C SC + + ++G+ G++ +V +
Sbjct 156 NVQVGDLIYGQFVVANKDMEPEM-----VCIDSCGR--ANGMGVIGQ--DGLLFKVTLGL 206
Query 274 ALTLLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCITA 323
LL D ++Q +G P EI G+NGR+W++A+T ++ +AN + A
Sbjct 207 IRKLLAPDCEIIQEVGKLYPLEIVFGMNGRIWVKAKTIQQTLILANILEA 256
> Hs7705606
Length=220
Score = 83.6 bits (205), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 50/167 (29%), Positives = 91/167 (54%), Gaps = 14/167 (8%)
Query 161 YVCISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLS 220
Y S RY P GD V+GI+++K + + V++ PA L+ ++ FEGAT+R++P +
Sbjct 45 YWVDSQQKRYVPVKGDHVIGIVTAKSGDIFKVDVGGSEPASLSYLS-FEGATKRNRPNVQ 103
Query 221 VGSVL----LVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALT 276
VG ++ +V + P + C SC + + ++G+ G++ +V +
Sbjct 104 VGDLIYGQFVVANKDMEPEM-----VCIDSCGR--ANGMGVIGQ--DGLLFKVTLGLIRK 154
Query 277 LLCHDSLVLQLLGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCITA 323
LL D ++Q +G P EI G+NGR+W++A+T ++ +AN + A
Sbjct 155 LLAPDCEIIQEVGKLYPLEIVFGMNGRIWVKAKTIQQTLILANILEA 201
> At2g25350_2
Length=157
Score = 60.8 bits (146), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 49/154 (31%), Positives = 68/154 (44%), Gaps = 24/154 (15%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGSVLLVR 228
Y PR D V+GI+ E Y ++I+ A L +A FEGA RR+ P+ + LL
Sbjct 3 EYVPRPEDHVLGIVVDCKGENYWIDIKGPQLALLPVLA-FEGANRRNYPKFE--AKLLDN 59
Query 229 VSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSL-VLQL 287
V G G L+ G + E + + LL + VL+
Sbjct 60 VIESGKAAG--------------------FGPLKDGFMFETSTGLSRMLLSSPTCPVLEA 99
Query 288 LGSRLPFEIAVGLNGRVWLRAETPTFSMRVANCI 321
LG +L FE A GLNGR W+ A P + VAN +
Sbjct 100 LGKKLSFETAFGLNGRCWVHAAAPRIVIIVANAL 133
> ECU09g1240
Length=189
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 49/151 (32%), Positives = 78/151 (51%), Gaps = 22/151 (14%)
Query 164 ISTNNRYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLATVAGFEGATRRSKPQLSVGS 223
+S NRY P + DIV+G + + + V++ A L +++ F AT+R+KP++S G
Sbjct 45 VSKTNRYQPCIDDIVIGKVVLTSQDMHKVDLGGTVGA-LPSLS-FTNATKRNKPEVSKGD 102
Query 224 VLLVRVSHLS--PLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHD 281
LL ++ + PLL TC G+L+G V+ P+ + LL +
Sbjct 103 HLLCKIVRMGTEPLL----TCVGEG-----------FGKLKGAVLP--LSPWKVRLL-YL 144
Query 282 SLVLQLLGSRLPFEIAVGLNGRVWLRAETPT 312
S L ++G + F IA+GLNG VW+ AE T
Sbjct 145 SDTLSIVGKKYRFRIALGLNGFVWIDAEKDT 175
> YHR069c
Length=359
Score = 37.4 bits (85), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 43/145 (29%), Positives = 61/145 (42%), Gaps = 19/145 (13%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGVNIRSVGPA--RLATVAGFEGATRRS--KPQLSVGSV 224
RY P GD VVG I+ G++ + V+I A L +V G RR +L + S
Sbjct 103 RYAPETGDHVVGRIAEVGNKRWKVDIGGKQHAVLMLGSVNLPGGILRRKSESDELQMRSF 162
Query 225 LLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLV 284
L LL A+V S + G+LR G+ +V P +L + +
Sbjct 163 L-----KEGDLLNAEVQSLFQDGSASLHTRSLKYGKLRNGMFCQV--PSSLIVRAKNHT- 214
Query 285 LQLLGSRLPFEIAV--GLNGRVWLR 307
LP I V G+NG +WLR
Sbjct 215 -----HNLPGNITVVLGVNGYIWLR 234
> At1g03360
Length=322
Score = 35.4 bits (80), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 36/142 (25%), Positives = 66/142 (46%), Gaps = 14/142 (9%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGV--NIRSVGPARLATVAGFEGATRR--SKPQLSVGSV 224
RY P VGDIVVG + + + V N G L+++ +G RR S +L++ ++
Sbjct 88 RYKPEVGDIVVGRVIEVAQKRWRVELNFNQDGVLMLSSMNMPDGIQRRRTSVDELNMRNI 147
Query 225 LLVRVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLV 284
V H ++ A+V S + + G+L G +++V P+ + H
Sbjct 148 F---VEH--DVVCAEVRNFQHDGSLQLQARSQKYGKLEKGQLLKVD-PYLVKRSKHHFHY 201
Query 285 LQLLGSRLPFEIAVGLNGRVWL 306
++ LG ++ +G NG +W+
Sbjct 202 VESLG----IDLIIGCNGFIWV 219
> SPAC2F7.14c
Length=329
Score = 32.3 bits (72), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 37/139 (26%), Positives = 61/139 (43%), Gaps = 9/139 (6%)
Query 169 RYCPRVGDIVVGIISSKGSEYYGVNIRSVGPARLA-TVAGFEGATRRSKPQLSVGSVLLV 227
+Y P +GD+++G I+ + + V+I + A L + G +R K L + +
Sbjct 106 KYVPEIGDLIIGKIAEVQPKRWKVDIGAKQNAVLMLSSINLPGGIQRRK--LETDELQMR 163
Query 228 RVSHLSPLLGADVTCCSSSCSKPWTSNEKLLGELRGGVIVEVAIPFALTLLCHDSLVLQL 287
LL A+V S S + G+LR GV ++V P AL + S L
Sbjct 164 SFFQEGDLLVAEVQQYFSDGSVSIHTRSLKYGKLRNGVFLKV--PPALVVRSK-SHAYAL 220
Query 288 LGSRLPFEIAVGLNGRVWL 306
G +I + +NG VW+
Sbjct 221 AGG---VDIILSVNGYVWV 236
> YOL084w
Length=991
Score = 32.0 bits (71), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/56 (30%), Positives = 25/56 (44%), Gaps = 1/56 (1%)
Query 102 RSFQWSAVY-VHPSHPSGKDETQNHGPESAVEVEEAGPCVKARTASMMLPKHVLPP 156
R F +S V + P +D+ HGPE AV V + + A++M PP
Sbjct 822 RKFHYSDVEALRNKRPYDEDDHSKHGPEGAVPVNADAGVIYSDPAAVMKEPQAFPP 877
> At3g19553
Length=479
Score = 30.4 bits (67), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 37/69 (53%), Gaps = 2/69 (2%)
Query 201 RLATVAGFEGATRRSKPQLSVGSVLLVRVSHLSPLLGADVTCCSSSCSKPWTSNE-KLLG 259
+ +T+AG ++ P+ G+VLLV S+L PL+ A SSS S W+ +G
Sbjct 218 KASTLAGEVDRPGKTFPKALFGAVLLVMGSYLIPLM-AGTGALSSSTSGEWSDGYFAEVG 276
Query 260 ELRGGVIVE 268
L GGV ++
Sbjct 277 MLIGGVWLK 285
Lambda K H
0.320 0.133 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8234976204
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40