bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3379_orf1
Length=206
Score E
Sequences producing significant alignments: (Bits) Value
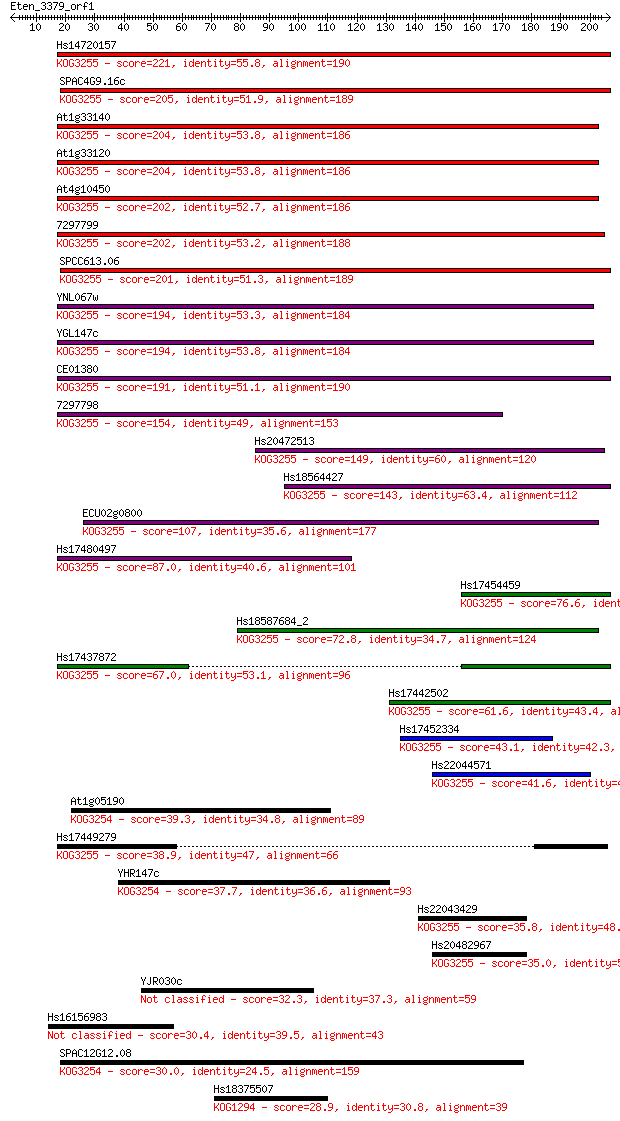
Hs14720157 221 1e-57
SPAC4G9.16c 205 6e-53
At1g33140 204 1e-52
At1g33120 204 1e-52
At4g10450 202 3e-52
7297799 202 4e-52
SPCC613.06 201 8e-52
YNL067w 194 7e-50
YGL147c 194 8e-50
CE01380 191 7e-49
7297798 154 1e-37
Hs20472513 149 4e-36
Hs18564427 143 2e-34
ECU02g0800 107 2e-23
Hs17480497 87.0 3e-17
Hs17454459 76.6 3e-14
Hs18587684_2 72.8 5e-13
Hs17437872 67.0 2e-11
Hs17442502 61.6 1e-09
Hs17452334 43.1 4e-04
Hs22044571 41.6 0.001
At1g05190 39.3 0.006
Hs17449279 38.9 0.007
YHR147c 37.7 0.017
Hs22043429 35.8 0.060
Hs20482967 35.0 0.095
YJR030c 32.3 0.76
Hs16156983 30.4 2.7
SPAC12G12.08 30.0 3.4
Hs18375507 28.9 8.4
> Hs14720157
Length=192
Score = 221 bits (562), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 106/191 (55%), Positives = 138/191 (72%), Gaps = 1/191 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHT-TKDAKRVKVATW 75
MKT+ S Q+V +P+ V ++++ R V VKGP G L+R F H+ + L K KR++V W
Sbjct 1 MKTILSNQTVDIPENVDITLKGRTVIVKGPRGTLRRDFNHINVELSLLGKKKKRLRVDKW 60
Query 76 QGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGE 135
G + ++A +RT+CSH+ NM GVT F YKMR VY+HFPIN+ I E G +VEIRNFLGE
Sbjct 61 WGNRKELATVRTICSHVQNMIKGVTLGFRYKMRSVYAHFPINVVIQENGSLVEIRNFLGE 120
Query 136 KRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIY 195
K +R V++ PGV C S KDE+ L+G D+ELVS SAALI QAT V+ KDIRKFLDGIY
Sbjct 121 KYIRRVRMRPGVACSVSQAQKDELILEGNDIELVSNSAALIQQATTVKNKDIRKFLDGIY 180
Query 196 VSEKGTLEKTD 206
VSEKGT+++ D
Sbjct 181 VSEKGTVQQAD 191
> SPAC4G9.16c
Length=190
Score = 205 bits (521), Expect = 6e-53, Method: Compositional matrix adjust.
Identities = 98/189 (51%), Positives = 131/189 (69%), Gaps = 2/189 (1%)
Query 18 KTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQG 77
+ +Y ++++ +P GVTV +++R VTV GP G LK+ RH+ + + K +K W G
Sbjct 3 RDIYKDETLTIPKGVTVDIKARNVTVTGPRGTLKQNLRHVDIEMK--KQGNTIKFIVWHG 60
Query 78 KKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKR 137
+ AC+R+V S +NNM GVT+ F YKMR VY+HFPIN+N+ E G VVEIRNFLGE+
Sbjct 61 SRKHNACIRSVYSIINNMIIGVTQGFRYKMRLVYAHFPININLTENGTVVEIRNFLGERI 120
Query 138 VRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVS 197
R++K LPGV S+ VKDEI L+G LE VS+SAA I Q VR KDIRKFLDGIYVS
Sbjct 121 TRVIKCLPGVTVSISSAVKDEIILEGNSLENVSQSAANIKQICNVRNKDIRKFLDGIYVS 180
Query 198 EKGTLEKTD 206
E+G +E+ +
Sbjct 181 ERGNIEELE 189
> At1g33140
Length=194
Score = 204 bits (518), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 100/190 (52%), Positives = 135/190 (71%), Gaps = 4/190 (2%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDA----KRVKV 72
MKT+ S +++ +PD VT+ V ++ + V+GP G+L R F+HL L KD K++K+
Sbjct 1 MKTILSSETMDIPDSVTIKVHAKVIEVEGPRGKLVRDFKHLNLDFQLIKDPETGKKKLKI 60
Query 73 ATWQGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNF 132
+W G + A +RT SH++N+ +GVT+ F YKMRFVY+HFPIN +I GK +EIRNF
Sbjct 61 DSWFGTRKTSASIRTALSHVDNLISGVTRGFRYKMRFVYAHFPINASIGGDGKSIEIRNF 120
Query 133 LGEKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLD 192
LGEK+VR V++L GV +S VKDEI L G D+ELVSRS ALI+Q V++KDIRKFLD
Sbjct 121 LGEKKVRKVEMLDGVTIVRSEKVKDEIVLDGNDIELVSRSCALINQKCHVKKKDIRKFLD 180
Query 193 GIYVSEKGTL 202
GIYVSEK +
Sbjct 181 GIYVSEKSKI 190
> At1g33120
Length=194
Score = 204 bits (518), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 100/190 (52%), Positives = 135/190 (71%), Gaps = 4/190 (2%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDA----KRVKV 72
MKT+ S +++ +PD VT+ V ++ + V+GP G+L R F+HL L KD K++K+
Sbjct 1 MKTILSSETMDIPDSVTIKVHAKVIEVEGPRGKLVRDFKHLNLDFQLIKDPETGKKKLKI 60
Query 73 ATWQGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNF 132
+W G + A +RT SH++N+ +GVT+ F YKMRFVY+HFPIN +I GK +EIRNF
Sbjct 61 DSWFGTRKTSASIRTALSHVDNLISGVTRGFRYKMRFVYAHFPINASIGGDGKSIEIRNF 120
Query 133 LGEKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLD 192
LGEK+VR V++L GV +S VKDEI L G D+ELVSRS ALI+Q V++KDIRKFLD
Sbjct 121 LGEKKVRKVEMLDGVTIVRSEKVKDEIVLDGNDIELVSRSCALINQKCHVKKKDIRKFLD 180
Query 193 GIYVSEKGTL 202
GIYVSEK +
Sbjct 181 GIYVSEKSKI 190
> At4g10450
Length=194
Score = 202 bits (515), Expect = 3e-52, Method: Compositional matrix adjust.
Identities = 98/190 (51%), Positives = 135/190 (71%), Gaps = 4/190 (2%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDA----KRVKV 72
MKT+ S +++ +PDGV + V ++ + V+GP G+L R F+HL L KD +++K+
Sbjct 1 MKTILSSETMDIPDGVAIKVNAKVIEVEGPRGKLTRDFKHLNLDFQLIKDQVTGKRQLKI 60
Query 73 ATWQGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNF 132
+W G + A +RT SH++N+ GVT+ F Y+MRFVY+HFPIN +I K +EIRNF
Sbjct 61 DSWFGSRKTSASIRTALSHVDNLIAGVTQGFLYRMRFVYAHFPINASIDGNNKSIEIRNF 120
Query 133 LGEKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLD 192
LGEK+VR V++L GV +S VKDEI L+G D+ELVSRS ALI+Q V++KDIRKFLD
Sbjct 121 LGEKKVRKVEMLDGVKIVRSEKVKDEIILEGNDIELVSRSCALINQKCHVKKKDIRKFLD 180
Query 193 GIYVSEKGTL 202
GIYVSEKG +
Sbjct 181 GIYVSEKGKI 190
> 7297799
Length=190
Score = 202 bits (514), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 100/188 (53%), Positives = 132/188 (70%), Gaps = 1/188 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQ 76
M+T+ S Q V++P + SV++R VT+ G G LKR+F+HL L ++ D + +KV W
Sbjct 1 MRTINSNQCVKIPKDIKASVKARVVTITGTRGTLKRSFKHLALDMYM-PDKRTLKVEKWF 59
Query 77 GKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEK 136
G K ++A +RTVCSH+ NM GVT F+YKMR VY+HFPIN E V+EIRNFLGEK
Sbjct 60 GTKKELAAVRTVCSHIENMIKGVTFGFQYKMRAVYAHFPINCVTSENNTVIEIRNFLGEK 119
Query 137 RVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYV 196
+R V++ PGV ST KDE+ ++G D+E VS SAALI Q+T V+ KDIRKFLDG+YV
Sbjct 120 YIRRVEMAPGVTVVNSTAQKDELIVEGNDIESVSGSAALIQQSTTVKNKDIRKFLDGLYV 179
Query 197 SEKGTLEK 204
SEK T+ K
Sbjct 180 SEKTTVVK 187
> SPCC613.06
Length=189
Score = 201 bits (511), Expect = 8e-52, Method: Compositional matrix adjust.
Identities = 97/189 (51%), Positives = 131/189 (69%), Gaps = 2/189 (1%)
Query 18 KTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQG 77
+ +Y ++++ +P+GV+V +++R VTVKGP G LK+ R + + L K +K W G
Sbjct 3 RDIYKDETLTIPEGVSVDIKARLVTVKGPRGVLKQNLRRVDIELK--KQGNTIKFIVWHG 60
Query 78 KKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKR 137
+ AC+RT S +NNM GVT+ F YKMR VY+HFPIN+N+ E G VVEIRNFLGE+
Sbjct 61 SRKHNACIRTAYSIINNMIIGVTQGFRYKMRLVYAHFPININLTENGTVVEIRNFLGERI 120
Query 138 VRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVS 197
R++K LPGV S+ VKDEI ++G LE VS+SAA I Q VR KDIRKFLDGIYVS
Sbjct 121 TRVIKCLPGVTVSISSAVKDEIIIEGNSLENVSQSAANIKQICNVRNKDIRKFLDGIYVS 180
Query 198 EKGTLEKTD 206
E+G +E+ +
Sbjct 181 ERGNIEELE 189
> YNL067w
Length=191
Score = 194 bits (494), Expect = 7e-50, Method: Compositional matrix adjust.
Identities = 98/186 (52%), Positives = 132/186 (70%), Gaps = 3/186 (1%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQ 76
MK + +EQ + +P+GVTVS++SR V V GP G L + +H+ + T + + +KVA
Sbjct 1 MKYIQTEQQIEIPEGVTVSIKSRIVKVVGPRGTLTKNLKHIDVTF-TKVNNQLIKVAVHN 59
Query 77 GKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIE--GGKVVEIRNFLG 134
G + +A LRTV S ++NM TGVTK ++YKMR+VY+HFPIN+NI+E G K +E+RNFLG
Sbjct 60 GDRKHVAALRTVKSLVDNMITGVTKGYKYKMRYVYAHFPINVNIVEKDGAKFIEVRNFLG 119
Query 135 EKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGI 194
+K++R V + GV E STNVKDEI L G +E VS++AA + Q VR KDIRKFLDGI
Sbjct 120 DKKIRNVPVRDGVTIEFSTNVKDEIVLSGNSVEDVSQNAADLQQICRVRNKDIRKFLDGI 179
Query 195 YVSEKG 200
YVS KG
Sbjct 180 YVSHKG 185
> YGL147c
Length=191
Score = 194 bits (494), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 99/186 (53%), Positives = 132/186 (70%), Gaps = 3/186 (1%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQ 76
MK + +EQ + VP+GVTVS++SR V V GP G L + +H+ + T + + +KVA
Sbjct 1 MKYIQTEQQIEVPEGVTVSIKSRIVKVVGPRGTLTKNLKHIDVTF-TKVNNQLIKVAVHN 59
Query 77 GKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIE--GGKVVEIRNFLG 134
G + +A LRTV S ++NM TGVTK ++YKMR+VY+HFPIN+NI+E G K +E+RNFLG
Sbjct 60 GGRKHVAALRTVKSLVDNMITGVTKGYKYKMRYVYAHFPINVNIVEKDGAKFIEVRNFLG 119
Query 135 EKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGI 194
+K++R V + GV E STNVKDEI L G +E VS++AA + Q VR KDIRKFLDGI
Sbjct 120 DKKIRNVPVRDGVTIEFSTNVKDEIVLSGNSVEDVSQNAADLQQICRVRNKDIRKFLDGI 179
Query 195 YVSEKG 200
YVS KG
Sbjct 180 YVSHKG 185
> CE01380
Length=189
Score = 191 bits (486), Expect = 7e-49, Method: Compositional matrix adjust.
Identities = 97/190 (51%), Positives = 131/190 (68%), Gaps = 1/190 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQ 76
MK + S +V P+GVT +V++R V V GP G +++ FRHL + + + ++V W
Sbjct 1 MKLIESNDTVVFPEGVTFTVKNRIVHVTGPRGTIRKDFRHLHMEMERIGKST-LRVRKWF 59
Query 77 GKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEK 136
G + ++A +RTVCSH+ NM GVT F YKMR VY+HFPIN+ + +G + VEIRNFLGEK
Sbjct 60 GVRKELAAIRTVCSHIKNMIKGVTVGFRYKMRSVYAHFPINVTLQDGNRTVEIRNFLGEK 119
Query 137 RVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYV 196
VR V L GV+ ST KDEI ++G D++ VS++AA I Q+T V+ KDIRKFLDGIYV
Sbjct 120 IVRRVPLPEGVIATISTAQKDEIVVEGNDVQFVSQAAARIQQSTAVKEKDIRKFLDGIYV 179
Query 197 SEKGTLEKTD 206
SEK T+ TD
Sbjct 180 SEKTTIVPTD 189
> 7297798
Length=208
Score = 154 bits (389), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 75/153 (49%), Positives = 103/153 (67%), Gaps = 1/153 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQ 76
M+T+ S Q V++P + SV++R VT+ G G LKR+F+HL L ++ D + +KV W
Sbjct 1 MRTINSNQCVKIPKDIKASVKARVVTITGTRGTLKRSFKHLALDMYMP-DKRTLKVEKWF 59
Query 77 GKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEK 136
G K ++A +RTVCSH+ NM GVT F+YKMR VY+HFPIN E V+EIRNFLGEK
Sbjct 60 GTKKELAAVRTVCSHIENMIKGVTFGFQYKMRAVYAHFPINCVTSENNTVIEIRNFLGEK 119
Query 137 RVRIVKLLPGVVCEKSTNVKDEIALKGTDLELV 169
+R V++ PGV ST KDE+ ++G D+E V
Sbjct 120 YIRRVEMAPGVTVVNSTAQKDELIVEGNDIESV 152
> Hs20472513
Length=125
Score = 149 bits (376), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 72/120 (60%), Positives = 87/120 (72%), Gaps = 0/120 (0%)
Query 85 LRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKRVRIVKLL 144
++T+CSH+ NM GVT F YKMR VY+HFPIN+ E G VEIRNFLGEK +R V++
Sbjct 2 VQTICSHVQNMIKGVTLGFHYKMRSVYAHFPINIVTRENGSFVEIRNFLGEKYIRRVRMR 61
Query 145 PGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEKGTLEK 204
PGV C K E+ L+G D+ELVS SAALI QAT V+ K IRKFLDGIYVSEK T ++
Sbjct 62 PGVACSVCQTQKHELILEGNDIELVSNSAALIQQATTVKNKHIRKFLDGIYVSEKETAQQ 121
> Hs18564427
Length=113
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 71/112 (63%), Positives = 85/112 (75%), Gaps = 0/112 (0%)
Query 95 MFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKRVRIVKLLPGVVCEKSTN 154
M GVT F YKMR VY+HFPIN+ I E G +VEIRNFLG+K +R V + GVVC S
Sbjct 1 MIKGVTLGFHYKMRSVYAHFPINVVIQENGSLVEIRNFLGKKYIRRVWMRAGVVCSVSQA 60
Query 155 VKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEKGTLEKTD 206
KDE+ L+G D+ELVS SAALI QAT V++KDIR+FLDGIYVSEKGT ++ D
Sbjct 61 QKDELILEGNDIELVSNSAALIQQATTVKKKDIREFLDGIYVSEKGTGQQAD 112
> ECU02g0800
Length=204
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 63/177 (35%), Positives = 95/177 (53%), Gaps = 3/177 (1%)
Query 26 VRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVATWQGKKSDIACL 85
V+VP+ T + + T +GP G + PL + R+++ W GK+ +A
Sbjct 30 VKVPEDCTATQEGKIFTFQGPKGSIVEDCTRHPLTFDIHEGNIRIRL--WNGKRKAMALA 87
Query 86 RTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKRVRIVKLLP 145
TV S L N VT F Y M+ VY+HF IN+ I E GK++ ++NFLGEK V+ + +
Sbjct 88 ITVESLLRNAIKAVTVGFAYTMKAVYNHFSINMEIKEDGKILLVKNFLGEKNVKRFR-MR 146
Query 146 GVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEKGTL 202
G + KD I ++G L VS+SA I + +D R FLDGI+V+E+G +
Sbjct 147 GAARVRLDERKDTIVIEGPSLSDVSQSAGTITNECKAKNRDSRVFLDGIFVTERGIM 203
> Hs17480497
Length=105
Score = 87.0 bits (214), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 41/102 (40%), Positives = 63/102 (61%), Gaps = 1/102 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHT-TKDAKRVKVATW 75
MKT+ S Q+V +P+ V ++++ V VKGP G L R F H+ + L K +R++V
Sbjct 1 MKTILSNQTVDIPENVDITLKGCTVIVKGPRGTLWRDFSHIHIELSLLGKKKQRLQVDKR 60
Query 76 QGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPIN 117
G + ++A ++ +CSH+ N GVT YKMR VY+HFP+N
Sbjct 61 WGNREELAAVQNICSHVQNTIKGVTLGLSYKMRSVYAHFPVN 102
> Hs17454459
Length=112
Score = 76.6 bits (187), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/51 (70%), Positives = 42/51 (82%), Gaps = 0/51 (0%)
Query 156 KDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEKGTLEKTD 206
KDE+ L+G D+ELVS S ALI QAT V+ KDIRKFLDGIYVSEKGT++ D
Sbjct 61 KDELILEGNDIELVSNSVALIQQATTVKIKDIRKFLDGIYVSEKGTVQPAD 111
> Hs18587684_2
Length=87
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 43/124 (34%), Positives = 62/124 (50%), Gaps = 37/124 (29%)
Query 79 KSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLGEKRV 138
+ ++A T+C+H+ N+ GVT F YKMR V++ FPINL I E G + +I+NFLGEK +
Sbjct 1 RKEVATACTICNHVQNIIKGVTLGFHYKMRSVHTPFPINLVIQEHGSLDKIQNFLGEKYI 60
Query 139 RIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSE 198
+ Q+T V+ KD RK D +YV+E
Sbjct 61 WM-------------------------------------QSTTVKNKDNRKLSDDLYVTE 83
Query 199 KGTL 202
KGT+
Sbjct 84 KGTV 87
> Hs17437872
Length=129
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 31/51 (60%), Positives = 41/51 (80%), Gaps = 0/51 (0%)
Query 156 KDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEKGTLEKTD 206
KDE+ L+G +++LVS SAA I QAT V+ KDIRK LDG YVSEKGT+++ +
Sbjct 57 KDELILEGNNIQLVSNSAASIQQATTVKNKDIRKILDGTYVSEKGTVQQAE 107
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 30/45 (66%), Gaps = 0/45 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVL 61
MKT+ S Q+V +P+ V ++++ R V VKGP G L R F H+ + L
Sbjct 1 MKTILSNQTVDIPENVDITLKGRTVIVKGPRGGLWRDFSHINVEL 45
> Hs17442502
Length=130
Score = 61.6 bits (148), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 33/81 (40%), Positives = 47/81 (58%), Gaps = 5/81 (6%)
Query 131 NFLGEKR-----VRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRK 185
+ LG++R + V++ PGV C S +DE+ L+ D+ELVS SA LI Q V+
Sbjct 33 SLLGKQRSSSGLTKWVQMRPGVACSVSQAKRDELILEENDVELVSSSAGLIQQVATVKTS 92
Query 186 DIRKFLDGIYVSEKGTLEKTD 206
RKF IYVS GT+++ D
Sbjct 93 KNRKFWGAIYVSVTGTVQQAD 113
> Hs17452334
Length=261
Score = 43.1 bits (100), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/52 (42%), Positives = 34/52 (65%), Gaps = 0/52 (0%)
Query 135 EKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKD 186
EK + V++ GV C S K+E+ L+ D+EL+S+S ALI QAT V+ ++
Sbjct 76 EKCIHKVQVRLGVACLISQTQKNELILEENDIELISKSVALIQQATTVKNRN 127
> Hs22044571
Length=109
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 32/54 (59%), Gaps = 0/54 (0%)
Query 146 GVVCEKSTNVKDEIALKGTDLELVSRSAALIHQATLVRRKDIRKFLDGIYVSEK 199
G ++T++ +L+ L+ V +AA I Q V+ KDIRKFL G+ VSEK
Sbjct 50 GFGLAQATSLSLNQSLQAVALQAVDSTAAWIQQTITVKSKDIRKFLYGVNVSEK 103
> At1g05190
Length=223
Score = 39.3 bits (90), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/93 (33%), Positives = 49/93 (52%), Gaps = 10/93 (10%)
Query 22 SEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAK---RVKVATWQGK 78
+Q + VP VT+++ +++ VKGP GEL T+ P + TK+ RVK +
Sbjct 47 GKQPIAVPSNVTIALEGQDLKVKGPLGELALTY---PREVELTKEESGFLRVKKTVETRR 103
Query 79 KSDIACL-RTVCSHLNNMFTGVTKQFEYKMRFV 110
+ + L RT+ +NM GV+K FE K+ V
Sbjct 104 ANQMHGLFRTLT---DNMVVGVSKGFEKKLILV 133
> Hs17449279
Length=113
Score = 38.9 bits (89), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 24/41 (58%), Gaps = 0/41 (0%)
Query 17 MKTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHL 57
MKT+ Q+V +P+ V ++++ V KGP G L F H+
Sbjct 1 MKTILCNQTVHIPENVDITLKGYPVIAKGPRGTLWMDFNHI 41
Score = 36.6 bits (83), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 16/25 (64%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 181 LVRRKDIRKFLDGIYVSEKGTLEKT 205
L + KDIRK +DGIYVSEKGT+++
Sbjct 87 LNKNKDIRKCVDGIYVSEKGTVKQA 111
> YHR147c
Length=214
Score = 37.7 bits (86), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 34/96 (35%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Query 38 SREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKV-ATWQGK--KSDIACLRTVCSHLNN 94
S+ +TVKGP GEL +P LH KD K K+ T Q K + TV S +NN
Sbjct 53 SQNITVKGPKGELS---VEVPDFLHLDKDEKHGKINVTVQNSEDKHQRSMWGTVRSLINN 109
Query 95 MFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIR 130
GVT+ +RFV + + L GK V ++
Sbjct 110 HIIGVTEGHLAVLRFVGTGYRAQLE--NDGKFVNVK 143
> Hs22043429
Length=318
Score = 35.8 bits (81), Expect = 0.060, Method: Compositional matrix adjust.
Identities = 18/37 (48%), Positives = 24/37 (64%), Gaps = 0/37 (0%)
Query 141 VKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALIH 177
V++ GV C S K E+ L+G D+ELVS SAAL+
Sbjct 204 VQMRLGVTCLVSQAQKVEVVLEGNDMELVSNSAALVQ 240
> Hs20482967
Length=113
Score = 35.0 bits (79), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 17/32 (53%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 146 GVVCEKSTNVKDEIALKGTDLELVSRSAALIH 177
GV C S K E+ L+G D+ELVS SAAL+
Sbjct 4 GVTCLVSQAQKVEVVLEGNDMELVSNSAALVQ 35
> YJR030c
Length=745
Score = 32.3 bits (72), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 31/70 (44%), Gaps = 11/70 (15%)
Query 46 PHGE-LKRTFRHLPLVLHTTKDA-----KRVKVATWQGKK-----SDIACLRTVCSHLNN 94
PH + + T+ PL ++T+ A VKV+ W GK D+ CL + S N
Sbjct 545 PHDDNVDETYVSFPLSINTSGGAVYFENDSVKVSLWNGKSWVPLSKDMLCLSLILSGDNE 604
Query 95 MFTGVTKQFE 104
V K FE
Sbjct 605 TLLIVYKDFE 614
> Hs16156983
Length=1199
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 14 LLKMKTVYSE--QSVRVPDGVTVSVRSREVTVKGPHGELKRTFRH 56
L K T+ S+ +S RVPD T + S+E+ +KG GE ++ H
Sbjct 332 LFKNPTLQSQLSRSHRVPDSSTATTSSKEIYLKGIAGEDTKSPHH 376
> SPAC12G12.08
Length=213
Score = 30.0 bits (66), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 39/162 (24%), Positives = 69/162 (42%), Gaps = 12/162 (7%)
Query 18 KTVYSEQSVRVPDGVTVSVRSREVTVKGPHGELKRTFRHLPLVLHTTKDAKRVKVAT--- 74
++ ++ + VP + + ++ + GPHG L F + L L T D +V +
Sbjct 31 RSYVGKKEIIVPKNIQFDLEGEQLQITGPHGTLNMRFPNY-LNLSKTNDILKVGMDANIE 89
Query 75 WQGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRFVYSHFPINLNIIEGGKVVEIRNFLG 134
Q K A T + L N GVT ++ ++ V + +LN +G ++I G
Sbjct 90 KQATKKQRAMWGTTRAILANNVKGVTMYWQSIIKLVGIGYRTSLN--DGNIHLKI----G 143
Query 135 EKRVRIVKLLPGVVCEKSTNVKDEIALKGTDLELVSRSAALI 176
+V + V E T + L+G D + V++ AA I
Sbjct 144 YANDILVSIPTDVQVENPTPT--SLVLRGIDRQKVTQFAAKI 183
> Hs18375507
Length=518
Score = 28.9 bits (63), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query 71 KVATWQGKKSDIACLRTVCSHLNNMFTGVTKQFEYKMRF 109
K+ TW+GK+ + + C H + G ++ +KMRF
Sbjct 139 KIRTWEGKEKTLTLINVYCPHAD---PGRPERLVFKMRF 174
Lambda K H
0.321 0.136 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3694581778
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40