bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3392_orf2
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
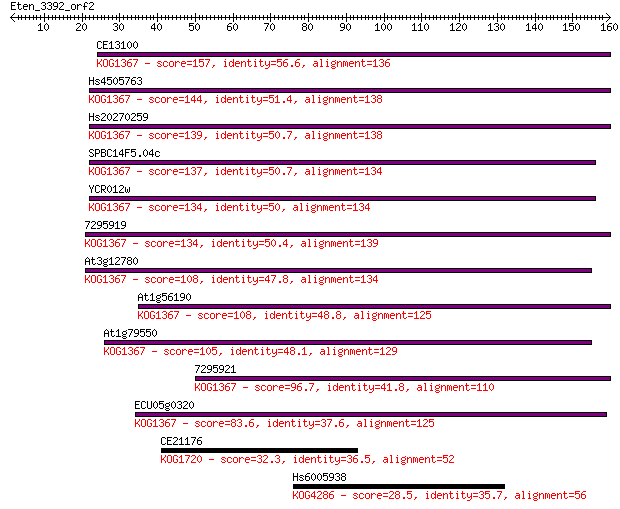
CE13100 157 6e-39
Hs4505763 144 9e-35
Hs20270259 139 3e-33
SPBC14F5.04c 137 1e-32
YCR012w 134 9e-32
7295919 134 1e-31
At3g12780 108 3e-24
At1g56190 108 6e-24
At1g79550 105 5e-23
7295921 96.7 2e-20
ECU05g0320 83.6 1e-16
CE21176 32.3 0.38
Hs6005938 28.5 5.3
> CE13100
Length=417
Score = 157 bits (398), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 77/136 (56%), Positives = 103/136 (75%), Gaps = 7/136 (5%)
Query 24 SKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLLLS 83
+K+ I+Q+ L GKRVL+RVDFNVPLKDGK+ + RIAA +PTI+ AL +GA++V+L+S
Sbjct 5 NKLAIDQL--NLAGKRVLIRVDFNVPLKDGKITNNQRIAAAVPTIQHALSNGAKSVVLMS 62
Query 84 HCGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEVLL 143
H GRPDGR Q +Y+L+PV L+ LL++ L F++DCVG + EAA A+ G V+L
Sbjct 63 HLGRPDGRRQDKYTLKPVAEELKALLKKDVL-----FLDDCVGSEVEAACADPAPGSVIL 117
Query 144 LENLRFHLEEEGKGED 159
LENLR+HLEEEGKG D
Sbjct 118 LENLRYHLEEEGKGVD 133
> Hs4505763
Length=417
Score = 144 bits (362), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 71/139 (51%), Positives = 102/139 (73%), Gaps = 8/139 (5%)
Query 22 LASKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLL 81
L++K+ ++++ +KGKRV+MRVDFNVP+K+ ++ + RI A +P+IKF L +GA++V+L
Sbjct 3 LSNKLTLDKL--DVKGKRVVMRVDFNVPMKNNQITNNQRIKAAVPSIKFCLDNGAKSVVL 60
Query 82 LSHCGRPDG-RVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGE 140
+SH GRPDG + +YSL PV L+ LL + L F++DCVGP+ E A AN G
Sbjct 61 MSHLGRPDGVPMPDKYSLEPVAVELKSLLGKDVL-----FLKDCVGPEVEKACANPAAGS 115
Query 141 VLLLENLRFHLEEEGKGED 159
V+LLENLRFH+EEEGKG+D
Sbjct 116 VILLENLRFHVEEEGKGKD 134
> Hs20270259
Length=417
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 70/139 (50%), Positives = 100/139 (71%), Gaps = 8/139 (5%)
Query 22 LASKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLL 81
L+ K+ ++++ ++GKRV+MRVDFNVP+K ++ + RI A+IP+IK+ L +GA+AV+L
Sbjct 3 LSKKLTLDKL--DVRGKRVIMRVDFNVPMKKNQITNNQRIKASIPSIKYCLDNGAKAVVL 60
Query 82 LSHCGRPDG-RVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGE 140
+SH GRPDG + +YSL PV L+ LL + L F++DCVG + E A AN G
Sbjct 61 MSHLGRPDGVPMPDKYSLAPVAVELKSLLGKDVL-----FLKDCVGAEVEKACANPAPGS 115
Query 141 VLLLENLRFHLEEEGKGED 159
V+LLENLRFH+EEEGKG+D
Sbjct 116 VILLENLRFHVEEEGKGQD 134
> SPBC14F5.04c
Length=414
Score = 137 bits (344), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 68/134 (50%), Positives = 92/134 (68%), Gaps = 7/134 (5%)
Query 22 LASKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLL 81
L++K+ I V LKGK VL+RVDFNVPL ++ + RI +PTIK+AL+ +AV+L
Sbjct 3 LSTKLAITDV--DLKGKNVLIRVDFNVPLDGDRITNNARIVGALPTIKYALEQQPKAVIL 60
Query 82 LSHCGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEV 141
+SH GRP+G +YSL+PV L +LL + V F++DCVGP+ E A A+ GEV
Sbjct 61 MSHLGRPNGARVAKYSLKPVAAELSKLLGKP-----VKFLDDCVGPEVEKACKEAKGGEV 115
Query 142 LLLENLRFHLEEEG 155
+LLENLRFH+EEEG
Sbjct 116 ILLENLRFHIEEEG 129
> YCR012w
Length=416
Score = 134 bits (336), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 67/134 (50%), Positives = 89/134 (66%), Gaps = 7/134 (5%)
Query 22 LASKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLL 81
L+SK+ ++ + LK KRV +RVDFNVPL K+ RI A +PTIK+ L+H R V+L
Sbjct 3 LSSKLSVQDL--DLKDKRVFIRVDFNVPLDGKKITSNQRIVAALPTIKYVLEHHPRYVVL 60
Query 82 LSHCGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEV 141
SH GRP+G ++YSL PV LQ LL + V+F+ DCVGP+ EAA + G V
Sbjct 61 ASHLGRPNGERNEKYSLAPVAKELQSLLGKD-----VTFLNDCVGPEVEAAVKASAPGSV 115
Query 142 LLLENLRFHLEEEG 155
+LLENLR+H+EEEG
Sbjct 116 ILLENLRYHIEEEG 129
> 7295919
Length=415
Score = 134 bits (336), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 70/139 (50%), Positives = 92/139 (66%), Gaps = 7/139 (5%)
Query 21 MLASKVGIEQVADQLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVL 80
M +K+ IE + L GKRVLMRVDFNVP+K+GK+ RI A + +IK AL A++V+
Sbjct 1 MAFNKLSIENL--DLAGKRVLMRVDFNVPIKEGKITSNQRIVAALDSIKLALSKKAKSVV 58
Query 81 LLSHCGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGE 140
L+SH GRPDG +Y+L PV L+ LL Q V F+ DCVG + EAA + G
Sbjct 59 LMSHLGRPDGNKNIKYTLAPVAAELKTLLGQD-----VIFLSDCVGSEVEAACKDPAPGS 113
Query 141 VLLLENLRFHLEEEGKGED 159
V+LLEN+RF++EEEGKG D
Sbjct 114 VILLENVRFYVEEEGKGLD 132
> At3g12780
Length=481
Score = 108 bits (271), Expect = 3e-24, Method: Composition-based stats.
Identities = 64/135 (47%), Positives = 88/135 (65%), Gaps = 9/135 (6%)
Query 21 MLASKVGIEQVADQLKGKRVLMRVDFNVPLKDGK-VADATRIAATIPTIKFALQHGARAV 79
M VG AD LKGK+V +R D NVPL D + + D TRI A IPTIK+ +++GA+ V
Sbjct 77 MAKKSVGDLTSAD-LKGKKVFVRADLNVPLDDNQTITDDTRIRAAIPTIKYLIENGAK-V 134
Query 80 LLLSHCGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNG 139
+L +H GRP G V ++SL P+VP L +LL +V+ +DC+GP+ E+ A+ G
Sbjct 135 ILSTHLGRPKG-VTPKFSLAPLVPRLSELL-----GIEVTKADDCIGPEVESLVASLPEG 188
Query 140 EVLLLENLRFHLEEE 154
VLLLEN+RF+ EEE
Sbjct 189 GVLLLENVRFYKEEE 203
> At1g56190
Length=478
Score = 108 bits (269), Expect = 6e-24, Method: Composition-based stats.
Identities = 61/126 (48%), Positives = 82/126 (65%), Gaps = 8/126 (6%)
Query 35 LKGKRVLMRVDFNVPLKDGK-VADATRIAATIPTIKFALQHGARAVLLLSHCGRPDGRVQ 93
LKGK+V +R D NVPL D + + D TRI A IPTIKF +++GA+ V+L +H GRP G V
Sbjct 87 LKGKKVFVRADLNVPLDDNQNITDDTRIRAAIPTIKFLIENGAK-VILSTHLGRPKG-VT 144
Query 94 QQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEVLLLENLRFHLEE 153
++SL P+VP L +LL +V +DC+GP+ E A+ G VLLLEN+RF+ EE
Sbjct 145 PKFSLAPLVPRLSELL-----GIEVVKADDCIGPEVETLVASLPEGGVLLLENVRFYKEE 199
Query 154 EGKGED 159
E D
Sbjct 200 EKNEPD 205
> At1g79550
Length=401
Score = 105 bits (261), Expect = 5e-23, Method: Compositional matrix adjust.
Identities = 62/130 (47%), Positives = 81/130 (62%), Gaps = 9/130 (6%)
Query 26 VGIEQVADQLKGKRVLMRVDFNVPLKD-GKVADATRIAATIPTIKFALQHGARAVLLLSH 84
VG + AD LKGK V +RVD NVPL D + D TRI A +PTIK+ + +G+R V+L SH
Sbjct 7 VGTLKEAD-LKGKSVFVRVDLNVPLDDNSNITDDTRIRAAVPTIKYLMGNGSR-VVLCSH 64
Query 85 CGRPDGRVQQQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEVLLL 144
GRP G V +YSL+P+VP L +LL + V D +G + + A G VLLL
Sbjct 65 LGRPKG-VTPKYSLKPLVPRLSELLGVE-----VVMANDSIGEEVQKLVAGLPEGGVLLL 118
Query 145 ENLRFHLEEE 154
EN+RF+ EEE
Sbjct 119 ENVRFYAEEE 128
> 7295921
Length=389
Score = 96.7 bits (239), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 46/110 (41%), Positives = 69/110 (62%), Gaps = 5/110 (4%)
Query 50 LKDGKVADATRIAATIPTIKFALQHGARAVLLLSHCGRPDGRVQQQYSLRPVVPVLQQLL 109
+KDGK+ + RI + +P+IK+AL ++++L SH GRPDG+ +++SL PV L+ +L
Sbjct 1 MKDGKITNNQRIVSALPSIKYALDKKCKSLVLASHLGRPDGKKNKKFSLEPVAKELESVL 60
Query 110 QQQQLSSKVSFVEDCVGPQAEAAAANAQNGEVLLLENLRFHLEEEGKGED 159
Q V F++DCVG A + +G V LLENLRF+ EE G +D
Sbjct 61 GQP-----VQFLDDCVGDSTLRALKDPSDGTVFLLENLRFYAEETGSSKD 105
> ECU05g0320
Length=388
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 47/125 (37%), Positives = 72/125 (57%), Gaps = 9/125 (7%)
Query 34 QLKGKRVLMRVDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLLLSHCGRPDGRVQ 93
+++GKRV +RVDFNVP+ D V + +I + +PTI+ R V++ SH GRP G+
Sbjct 8 EIEGKRVFLRVDFNVPILDSVVLNDFKITSVLPTIRLLHSRNPRRVIIGSHLGRPKGKYS 67
Query 94 QQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEAAAANAQNGEVLLLENLRFHLEE 153
++ SLRPV VL+ L ++L ++ F + N Q+ +LLENLRF+ E
Sbjct 68 KELSLRPVYSVLKARL-WEELGVELDFCDLW-------DVGNVQS-PWILLENLRFYSAE 118
Query 154 EGKGE 158
E G+
Sbjct 119 EDAGD 123
> CE21176
Length=446
Score = 32.3 bits (72), Expect = 0.38, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 7/55 (12%)
Query 41 LMR---VDFNVPLKDGKVADATRIAATIPTIKFALQHGARAVLLLSHCGRPDGRV 92
LMR +N PL D + R+ + + FAL HG AV HC GR
Sbjct 134 LMRNGIYHYNFPLPDFQACTPNRLLDIVKVVDFALSHGKIAV----HCHAGHGRT 184
> Hs6005938
Length=3433
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 4/60 (6%)
Query 76 ARAVLLLSHCGRPDGRVQ----QQYSLRPVVPVLQQLLQQQQLSSKVSFVEDCVGPQAEA 131
A A LL H GR + R+Q L + L+QLL+Q + S+++ V PQ A
Sbjct 3316 AEAKLLRQHKGRLEARMQILEDHNKQLESQLHRLRQLLEQPESDSRINGVSPWASPQHSA 3375
Lambda K H
0.321 0.135 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40