bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3400_orf1
Length=173
Score E
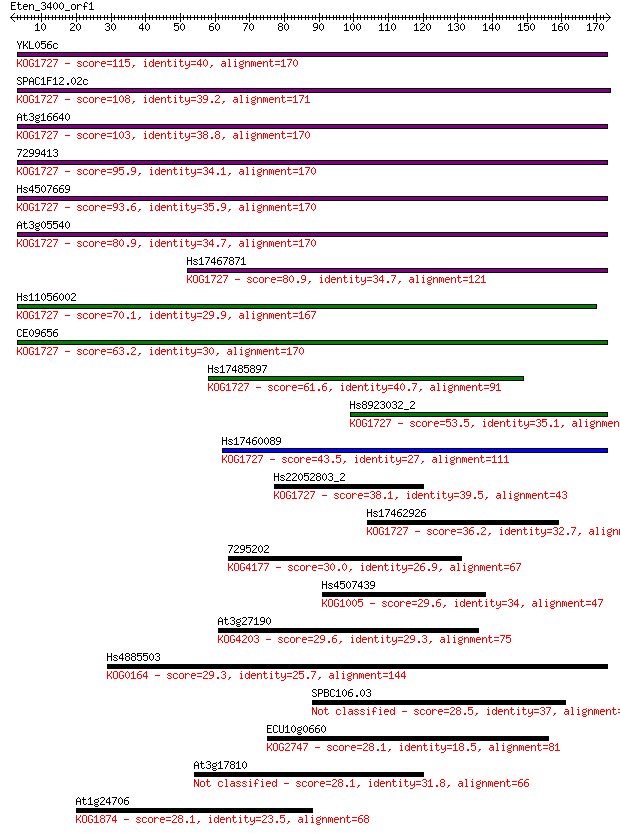
Sequences producing significant alignments: (Bits) Value
YKL056c 115 3e-26
SPAC1F12.02c 108 4e-24
At3g16640 103 2e-22
7299413 95.9 3e-20
Hs4507669 93.6 2e-19
At3g05540 80.9 1e-15
Hs17467871 80.9 1e-15
Hs11056002 70.1 2e-12
CE09656 63.2 3e-10
Hs17485897 61.6 7e-10
Hs8923032_2 53.5 2e-07
Hs17460089 43.5 2e-04
Hs22052803_2 38.1 0.010
Hs17462926 36.2 0.031
7295202 30.0 2.7
Hs4507439 29.6 2.8
At3g27190 29.6 3.1
Hs4885503 29.3 4.1
SPBC106.03 28.5 7.7
ECU10g0660 28.1 8.7
At3g17810 28.1 9.3
At1g24706 28.1 9.9
> YKL056c
Length=167
Score = 115 bits (289), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 68/172 (39%), Positives = 97/172 (56%), Gaps = 8/172 (4%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAEDFGIADN--SEEGE 60
M +YK +FS DEL SD+Y D DV +E V ++ I N +E G+
Sbjct 1 MIIYKDIFSNDELLSDAY------DAKLVDDVIYEADCAMVNVGGDNIDIGANPSAEGGD 54
Query 61 GGVEDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESA 120
VE+ AE V +++++F L +T+ KK F TYIKGY+K V LQE NP V F+ +
Sbjct 55 DDVEEGAEMVNNVVHSFRLQQTAFDKKSFLTYIKGYMKAVKAKLQETNPEEVPKFEKGAQ 114
Query 121 ALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEEK 172
VKK++GSF D++FF ES D +A +V Y+++ T P K G+ EEK
Sbjct 115 TYVKKVIGSFKDWEFFTGESMDPDAMVVMLNYREDGTTPFVAIWKHGIVEEK 166
> SPAC1F12.02c
Length=168
Score = 108 bits (271), Expect = 4e-24, Method: Compositional matrix adjust.
Identities = 67/174 (38%), Positives = 93/174 (53%), Gaps = 9/174 (5%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVT-KAAEDFGIADNSEEGEG 61
M +YK V SGDEL SD+Y D D+ +E VT K D I N +
Sbjct 1 MLLYKDVISGDELVSDAY------DLKEVDDIVYEADCQMVTVKQGGDVDIGANPSAEDA 54
Query 62 GVED--AAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTES 119
ETV +++ +F LS TS KK + +YIKGY+K + LQE NP RV F+ +
Sbjct 55 EENAEEGTETVNNLVYSFRLSPTSFDKKSYMSYIKGYMKAIKARLQESNPERVPVFEKNA 114
Query 120 AALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEEKY 173
VKKIL +F D+ F++ ES D +A +V Y+++ P I+ KDGL EK+
Sbjct 115 IGFVKKILANFKDYDFYIGESMDPDAMVVLMNYREDGITPYMIFFKDGLVSEKF 168
> At3g16640
Length=168
Score = 103 bits (257), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 66/175 (37%), Positives = 94/175 (53%), Gaps = 13/175 (7%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAEDFGIADNSEEGEGG 62
M VY+ + +GDEL SDS+P+ + + +EV+G VT A D I N EGG
Sbjct 1 MLVYQDLLTGDELLSDSFPY-----KEIENGILWEVEGKWVTVGAVDVNIGANPSAEEGG 55
Query 63 ----VEDAAETVIDIINAFHLSET-SLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKT 117
V+D+ + V+DI++ F L E + KK F YIK Y+K + L E + + FK
Sbjct 56 EDEGVDDSTQKVVDIVDTFRLQEQPTYDKKGFIAYIKKYIKLLTPKLSEEDQA---VFKK 112
Query 118 ESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEEK 172
K +L DFQFF+ E +++LV+AYYK+ T P F+Y GLKE K
Sbjct 113 GIEGATKFLLPRLSDFQFFVGEGMHDDSTLVFAYYKEGSTNPTFLYFAHGLKEVK 167
> 7299413
Length=172
Score = 95.9 bits (237), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 58/177 (32%), Positives = 99/177 (55%), Gaps = 13/177 (7%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAEDF---GIADNSEEG 59
MK+YK + +GDE+ +D+Y + DV +EV G +T+ +D G ++EE
Sbjct 1 MKIYKDIITGDEMFADTYKMKL------VDDVIYEVYGKLITRQGDDIKLEGANASAEEA 54
Query 60 EGGVEDAAETVIDIINAFHLSETSL--TKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKT 117
+ G + +E+ +D++ L+E KK +T Y+K Y+K+V+ L+E +P +V+ FKT
Sbjct 55 DEGTDITSESGVDVVLNHRLTECFAFGDKKSYTLYLKDYMKKVLAKLEEKSPDQVDIFKT 114
Query 118 ESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYK--DEETAPRFIYLKDGLKEEK 172
+K ILG F + QFF ES D + + Y+ + ++ P ++ K GL+EEK
Sbjct 115 NMNKAMKDILGRFKELQFFTGESMDCDGMVALVEYREINGDSVPVLMFFKHGLEEEK 171
> Hs4507669
Length=172
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 61/177 (34%), Positives = 94/177 (53%), Gaps = 13/177 (7%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKA---AEDFGIADNSE-- 57
M +Y+ + S DE+ SD Y D + EV+G V++ +D I N+
Sbjct 1 MIIYRDLISHDEMFSDIYKIREIADG-----LCLEVEGKMVSRTEGNIDDSLIGGNASAE 55
Query 58 --EGEGGVEDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAF 115
EGEG E T +DI+ HL ETS TK+ + YIK Y+K + L+E P RV+ F
Sbjct 56 GPEGEG-TESTVITGVDIVMNHHLQETSFTKEAYKKYIKDYMKSIKGKLEEQRPERVKPF 114
Query 116 KTESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEEK 172
T +A +K IL +F ++QFF+ E+ + + + Y+++ P I+ KDGL+ EK
Sbjct 115 MTGAAEQIKHILANFKNYQFFIGENMNPDGMVALLDYREDGVTPYMIFFKDGLEMEK 171
> At3g05540
Length=156
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 59/172 (34%), Positives = 87/172 (50%), Gaps = 19/172 (11%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAEDFGIADNSEEGEGG 62
M VY+ + +GDEL SDS+P+ + + +EV+G + + +
Sbjct 1 MLVYQDILTGDELLSDSFPY-----KEIENGMLWEVEGKNPSGEEGGEDEGVDDQ----- 50
Query 63 VEDAAETVIDIINAFHLSET-SLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESAA 121
A V+DII+ F L E S KK F ++K Y+K++ L N E FK +
Sbjct 51 ----AVKVVDIIDTFRLQEQPSFDKKQFVMFMKRYIKQLSPKLDSENQ---ELFKKHIES 103
Query 122 LVKKILGSFDDFQFFMSESNDFE-ASLVYAYYKDEETAPRFIYLKDGLKEEK 172
K ++ DFQFF+ ES + E SLV+AYY++ T P F+YL GLKE K
Sbjct 104 ATKFLMSKLKDFQFFVGESMEGEEGSLVFAYYREGATDPTFLYLAYGLKEIK 155
> Hs17467871
Length=138
Score = 80.9 bits (198), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 42/121 (34%), Positives = 67/121 (55%), Gaps = 1/121 (0%)
Query 52 IADNSEEGEGGVEDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSR 111
++ EGEG E T +D++ HL ETS TK+ YIK Y+K + L+E P R
Sbjct 18 VSAEGPEGEG-TESTVITAVDVVMNHHLQETSFTKEAHKKYIKDYMKSIKGKLEELRPER 76
Query 112 VEAFKTESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEE 171
V+ F T +A +K IL +F ++ FF+ E+ + + + Y ++ P I+ +DGL+ E
Sbjct 77 VKPFMTGAAEQIKHILANFKNYHFFIGENMNPDGMVALLDYGEDGVTPYMIFFRDGLETE 136
Query 172 K 172
K
Sbjct 137 K 137
> Hs11056002
Length=173
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 50/174 (28%), Positives = 87/174 (50%), Gaps = 12/174 (6%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAED-----FGIADNSE 57
M +++ + S +E+ SD Y + + EV+ ++K + G +SE
Sbjct 1 MIIFQDLISHNEMFSDIYKIW-----EITNGLCLEVEQKMLSKTTGNTDDSLIGRNSSSE 55
Query 58 EGEGGV-EDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFK 116
E V E T +DI+ HL E+ TK+ + YIK Y+K + + L+E P RV+ F
Sbjct 56 STEDEVTESTIITSVDIVTNHHLQESIFTKEAYKKYIKDYMKSINEKLEEQRPERVKLFI 115
Query 117 TESAALVKKILGSFDDFQFFMSES-NDFEASLVYAYYKDEETAPRFIYLKDGLK 169
T + +K I +F ++QFF+ E+ N + Y KD + P + +L++GL+
Sbjct 116 TGNEEQIKHIFANFQNYQFFIGENLNPPGMVALLNYRKDGQYDPIYDFLRNGLE 169
> CE09656
Length=181
Score = 63.2 bits (152), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 51/186 (27%), Positives = 90/186 (48%), Gaps = 22/186 (11%)
Query 3 MKVYKCVFSGDELCSDSYPHLVPFDDDAFKDVAFEVKGTRVTKAAEDFGIADNSEEGEGG 62
M +YK +F+ DEL SDS+P + D+ +E KG V + + +A ++ E G
Sbjct 1 MLIYKDIFTDDELSSDSFPMKL------VDDLVYEFKGKHVVRKEGEIVLAGSNPSAEEG 54
Query 63 VED-----AAETVIDIINAFHLSETSLTKKD--FTTYIKGYLKRVVDYLQEHNPSR--VE 113
ED E IDI+ L E + + F YIK ++K V+D+++++N + V+
Sbjct 55 AEDDGSDEHVERGIDIVLNHKLVEMNCYEDASMFKAYIKKFMKNVIDHMEKNNRDKADVD 114
Query 114 AFKTESAALVKKILG--SFDDFQFFMSE-----SNDFEASLVYAYYKDEETAPRFIYLKD 166
AFK + V +L F + FF+ E + + + +++ D P + +K+
Sbjct 115 AFKKKIQGWVVSLLAKDRFKNLAFFIGERAAEGAENGQVAIIEYRDVDGTEVPTLMLVKE 174
Query 167 GLKEEK 172
+ EEK
Sbjct 175 AIIEEK 180
> Hs17485897
Length=144
Score = 61.6 bits (148), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 37/98 (37%), Positives = 51/98 (52%), Gaps = 7/98 (7%)
Query 58 EGEGGVEDAAETVID-------IINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPS 110
E EG + D E ID + A + ETS TK+ YIK Y+K + L+E P
Sbjct 7 EVEGKIVDKTEGNIDDSLVEMLPLKASRVKETSFTKEACKKYIKHYMKSIKGRLEEQKPE 66
Query 111 RVEAFKTESAALVKKILGSFDDFQFFMSESNDFEASLV 148
RV+AF T A K I +F ++QFF+ E+ + AS V
Sbjct 67 RVKAFMTGPAEQTKHIFANFKNYQFFIGENMNTNASFV 104
> Hs8923032_2
Length=77
Score = 53.5 bits (127), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/75 (34%), Positives = 45/75 (60%), Gaps = 1/75 (1%)
Query 99 RVVDYL-QEHNPSRVEAFKTESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEET 157
+V+ +L E P RV+ F T +A +K IL +F ++QFF+ E+ + + + Y+++
Sbjct 2 KVIKFLGDEQRPERVKPFMTGAAEQIKHILANFKNYQFFIGENMNPDGMVALLDYREDGV 61
Query 158 APRFIYLKDGLKEEK 172
P I+ KDGL+ EK
Sbjct 62 TPYMIFFKDGLEMEK 76
> Hs17460089
Length=168
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/111 (27%), Positives = 47/111 (42%), Gaps = 22/111 (19%)
Query 62 GVEDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESAA 121
G E + II H ETS K+ + YIK Y +A
Sbjct 79 GTESTVFDAVVIIMKHHSPETSSAKEAYRKYIKNY----------------------TAE 116
Query 122 LVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETAPRFIYLKDGLKEEK 172
+K IL +F ++QF + E + + L Y++++ P I+ KD L+ EK
Sbjct 117 QIKHILANFKNYQFIICEDMNPDGMLALLDYQEDDVTPYMIFFKDDLEMEK 167
> Hs22052803_2
Length=102
Score = 38.1 bits (87), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 77 FHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTES 119
+HL ETS TK + YIK Y+K + L+E P +V ++S
Sbjct 56 YHLQETSFTKNTYKNYIKDYMKTIKGKLEEQRPEKVYHVHSDS 98
> Hs17462926
Length=190
Score = 36.2 bits (82), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 30/55 (54%), Gaps = 0/55 (0%)
Query 104 LQEHNPSRVEAFKTESAALVKKILGSFDDFQFFMSESNDFEASLVYAYYKDEETA 158
L+E P RV+ F T +A +K IL +F + FF+ E+ + + + Y+ E A
Sbjct 113 LEEQRPERVKPFMTGAAEQIKHILANFKSYHFFIGENMNPDGMVAPQDYQIEVQA 167
> 7295202
Length=2443
Score = 30.0 bits (66), Expect = 2.7, Method: Composition-based stats.
Identities = 18/67 (26%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 64 EDAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESAALV 123
E AE V ++++ FH E +TK + T+ I ++ +V ++E S+V+ + + A V
Sbjct 2037 ETVAEKVTELLDTFHKIEEKVTKSEKTSEISSKVEELVK-IEEKPLSQVQPKEDKVAEKV 2095
Query 124 KKILGSF 130
+++ +F
Sbjct 2096 AEVIETF 2102
> Hs4507439
Length=1132
Score = 29.6 bits (65), Expect = 2.8, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 0/47 (0%)
Query 91 TYIKGYLKRVVDYLQEHNPSRVEAFKTESAALVKKILGSFDDFQFFM 137
T ++ Y+++ V +LQE +P R +S++L + G FD F FM
Sbjct 767 TDLQPYMRQFVAHLQETSPLRDAVVIEQSSSLNEASSGLFDVFLRFM 813
> At3g27190
Length=483
Score = 29.6 bits (65), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/76 (28%), Positives = 33/76 (43%), Gaps = 3/76 (3%)
Query 61 GGVEDAAETVID-IINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTES 119
GG TV D II H L +D ++ +G ++++QE+N +AF TE
Sbjct 69 GGTASGKTTVCDMIIQQLHDHRIVLVNQD--SFYRGLTSEELEHVQEYNFDHPDAFDTEQ 126
Query 120 AALVKKILGSFDDFQF 135
IL S +Q
Sbjct 127 LLHCVDILKSGQPYQI 142
> Hs4885503
Length=1043
Score = 29.3 bits (64), Expect = 4.1, Method: Composition-based stats.
Identities = 37/153 (24%), Positives = 63/153 (41%), Gaps = 25/153 (16%)
Query 29 DAFKDVAFEVKGTRVTKAAEDFG--IADNSEEGEGGV-----EDAAETVIDIINAFH--L 79
D F+ G R + + G + NSEE E + E A E V+ +N
Sbjct 281 DEFQASGIPASGIRDGRGVREIGEMVGLNSEEVERALCSRTMETAKEKVVTALNVMQAQY 340
Query 80 SETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESAALVKKILGSFDDFQFFMSE 139
+ +L K ++ R+ D++ ++ E KK++G D + F + E
Sbjct 341 ARDALAKNIYS--------RLFDWIVNRINESIKVGIGEK----KKVMGVLDIYGFEILE 388
Query 140 SNDFEASLVYAYYKDEETAPRFIYLKDGLKEEK 172
N FE ++ Y +E+ FI + LKEE+
Sbjct 389 DNSFEQFVI--NYCNEKLQQVFIEMT--LKEEQ 417
> SPBC106.03
Length=357
Score = 28.5 bits (62), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 36/83 (43%), Gaps = 15/83 (18%)
Query 88 DFTTYIKGYLKRVVDYLQE--HNPSRVEAFKTESAALVKKILGSF---DDFQFFMSESND 142
D + +K Y KR++DY E NP V T SA K LG F + + +
Sbjct 9 DLVSNVKDYGKRIIDYAHEKTSNPQPVTVHTTNSA---KGRLGEFLYQNRVSVGLGSAAL 65
Query 143 FEASLVYAYYK-----DEETAPR 160
F A L +YYK + APR
Sbjct 66 FLAGL--SYYKRYDYVRKRRAPR 86
> ECU10g0660
Length=377
Score = 28.1 bits (61), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 15/81 (18%), Positives = 37/81 (45%), Gaps = 4/81 (4%)
Query 75 NAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPSRVEAFKTESAALVKKILGSFDDFQ 134
N E L+ +Y +++ +V+YL +H+ + + E+ ++G+ ++
Sbjct 281 NVVSGPEKPLSDLGLLSYRAYWMEVIVEYLSKHDKASISEISRETYISEDDVIGTLCAYK 340
Query 135 FFMSESNDFEASLVYAYYKDE 155
E N+F ++ Y +D+
Sbjct 341 MLRMEGNEF----IFTYAEDQ 357
> At3g17810
Length=426
Score = 28.1 bits (61), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 5/69 (7%)
Query 54 DNSEEGEGGVE---DAAETVIDIINAFHLSETSLTKKDFTTYIKGYLKRVVDYLQEHNPS 110
D S G GGVE DAAE ++ N + T + + ++K + D++++HN S
Sbjct 311 DRSLSGIGGVETGYDAAEFILLGSNTVQVC-TGVMMHGYG-HVKTLCAELKDFMKQHNFS 368
Query 111 RVEAFKTES 119
+E F+ S
Sbjct 369 TIEEFRGHS 377
> At1g24706
Length=1705
Score = 28.1 bits (61), Expect = 9.9, Method: Composition-based stats.
Identities = 16/70 (22%), Positives = 35/70 (50%), Gaps = 4/70 (5%)
Query 20 YPHLVPFDDDAFKDVAFEVKGTRVTKAAEDFGIADNSEEGEGGVED--AAETVIDIINAF 77
Y HL+P D++ F+D + V + + A G + + G+ +ED + +D+ A
Sbjct 183 YAHLLPKDEEVFED--YNVSSAKRFEEANKIGKINLAATGKDLMEDEKQGDVTVDLFAAL 240
Query 78 HLSETSLTKK 87
+ ++T++
Sbjct 241 DMESEAVTER 250
Lambda K H
0.316 0.135 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40