bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3405_orf2
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
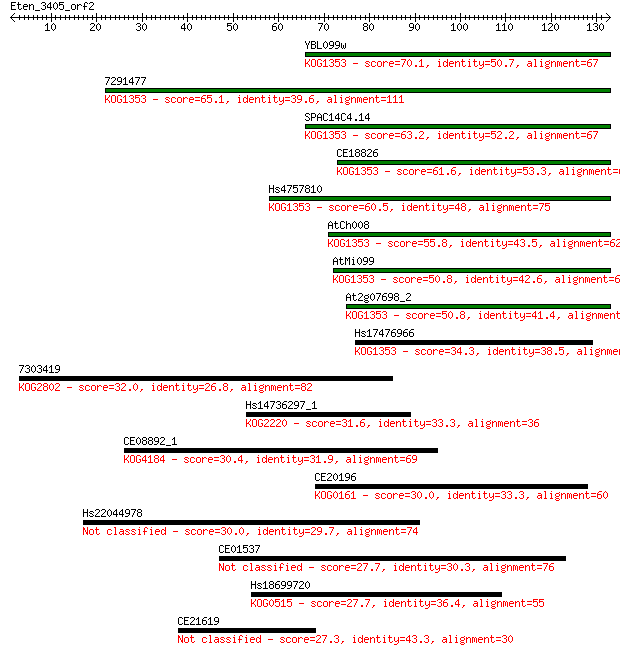
YBL099w 70.1 1e-12
7291477 65.1 4e-11
SPAC14C4.14 63.2 1e-10
CE18826 61.6 4e-10
Hs4757810 60.5 9e-10
AtCh008 55.8 2e-08
AtMi099 50.8 7e-07
At2g07698_2 50.8 8e-07
Hs17476966 34.3 0.065
7303419 32.0 0.33
Hs14736297_1 31.6 0.37
CE08892_1 30.4 1.0
CE20196 30.0 1.1
Hs22044978 30.0 1.3
CE01537 27.7 6.4
Hs18699720 27.7 6.7
CE21619 27.3 8.0
> YBL099w
Length=545
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 34/67 (50%), Positives = 46/67 (68%), Gaps = 1/67 (1%)
Query 66 RAAATTISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVEL 125
R A+T P+E++ ILE+RI G + ++ E GRV +VGDGIAR+ GL N++A ELVE
Sbjct 34 RLASTKAQPTEVSSILEERIKG-VSDEANLNETGRVLAVGDGIARVFGLNNIQAEELVEF 92
Query 126 QSGAVGM 132
SG GM
Sbjct 93 SSGVKGM 99
> 7291477
Length=552
Score = 65.1 bits (157), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 44/111 (39%), Positives = 61/111 (54%), Gaps = 7/111 (6%)
Query 22 LSSAAAACAARLSARLPAAAAAAAGPAAAAGQQQQRRLLHTSDLRAAATTISPSEMAKIL 81
+S +A A+ ++ LP AA A AA R LH A+T +E++ IL
Sbjct 1 MSIFSARLASSVARNLPKAANQVACKAAYPAASLAARKLHV------ASTQRSAEISNIL 54
Query 82 EQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVELQSGAVGM 132
E+RI G ++ E GRV S+GDGIAR+ GL N++A E+VE SG GM
Sbjct 55 EERILG-VAPKADLEETGRVLSIGDGIARVYGLNNIQADEMVEFSSGLKGM 104
> SPAC14C4.14
Length=536
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 43/67 (64%), Gaps = 1/67 (1%)
Query 66 RAAATTISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVEL 125
R A +P+E+ ILE+RI G Q+ E GRV S+GDGIAR+ GL NV+A ELVE
Sbjct 25 RGYAEKAAPTEVPSILEERIRG-AYNQAQMMESGRVLSIGDGIARISGLSNVQAEELVEF 83
Query 126 QSGAVGM 132
SG GM
Sbjct 84 SSGIKGM 90
> CE18826
Length=538
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 32/60 (53%), Positives = 42/60 (70%), Gaps = 1/60 (1%)
Query 73 SPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVELQSGAVGM 132
S SE++KILE+RI G T + E G+V S+GDGIAR+ GL N++A E+VE SG GM
Sbjct 32 SGSEVSKILEERILGTETGINLE-ETGKVLSIGDGIARVYGLKNIQAEEMVEFDSGIKGM 90
> Hs4757810
Length=553
Score = 60.5 bits (145), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 36/75 (48%), Positives = 44/75 (58%), Gaps = 3/75 (4%)
Query 58 RLLHTSDLRAAATTISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNV 117
R H S+ T +EM+ ILE+RI G T E GRV S+GDGIAR+ GL NV
Sbjct 34 RNFHASNTHLQKT--GTAEMSSILEERILGADTSVDLE-ETGRVLSIGDGIARVHGLRNV 90
Query 118 RAGELVELQSGAVGM 132
+A E+VE SG GM
Sbjct 91 QAEEMVEFSSGLKGM 105
> AtCh008
Length=507
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 27/62 (43%), Positives = 38/62 (61%), Gaps = 1/62 (1%)
Query 71 TISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVELQSGAV 130
TI E++ I+ +RI + ++ T G V VGDGIAR+ GL V AGELVE + G +
Sbjct 3 TIRADEISNIIRERIEQYN-REVTIVNTGTVLQVGDGIARIYGLDEVMAGELVEFEEGTI 61
Query 131 GM 132
G+
Sbjct 62 GI 63
> AtMi099
Length=507
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/63 (41%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query 72 ISP--SEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVELQSGA 129
+SP +E+ + E RI + E+GRV SVGDGIA++ GL ++AGE+V +G
Sbjct 3 LSPRAAELTNLFESRIRNFYAN-FQVDEIGRVVSVGDGIAQVYGLNEIQAGEMVLFANGV 61
Query 130 VGM 132
GM
Sbjct 62 KGM 64
> At2g07698_2
Length=507
Score = 50.8 bits (120), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 24/58 (41%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 75 SEMAKILEQRISGWTTQQSTAGEVGRVFSVGDGIARLLGLGNVRAGELVELQSGAVGM 132
+E+ + E RI + E+GRV SVGDGIA++ GL ++AGE+V +G GM
Sbjct 8 AELTNLFESRIRNFYAN-FQVDEIGRVVSVGDGIAQVYGLNEIQAGEMVLFANGVKGM 64
> Hs17476966
Length=172
Score = 34.3 bits (77), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 32/55 (58%), Gaps = 7/55 (12%)
Query 77 MAKILEQRISGWTTQQSTAGEVGR---VFSVGDGIARLLGLGNVRAGELVELQSG 128
M+ I E+ I + +T+ ++ V S+G GIA++ GL NV+A E+ E SG
Sbjct 1 MSSIFEEHI----VRDNTSADLEETQPVLSIGGGIAQVHGLTNVQAEEVEEFSSG 51
> 7303419
Length=655
Score = 32.0 bits (71), Expect = 0.33, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 37/85 (43%), Gaps = 3/85 (3%)
Query 3 RPRRRSFVSILKMAPLIRPLSSAAAACAARLS---ARLPAAAAAAAGPAAAAGQQQQRRL 59
P RR K+ P RP + C+A S P A AGP A + + +
Sbjct 31 NPWRRHDGEAFKLQPWKRPSTFLQFRCSANGSDSKKEKPVQEEATAGPVAKPKETKNFEV 90
Query 60 LHTSDLRAAATTISPSEMAKILEQR 84
T + + +TTI S++ +I+ ++
Sbjct 91 KTTKGILSISTTIEDSKINEIVFEK 115
> Hs14736297_1
Length=1189
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 53 QQQQRRLLHTSDLRAAATTISPSEMAKILEQRISGW 88
+QQ R L+ D+ A+ T SEM K+ E+++ +
Sbjct 574 EQQLRELIQKDDITASLVTTDHSEMKKLFEEQLKKY 609
> CE08892_1
Length=452
Score = 30.4 bits (67), Expect = 1.0, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 30/73 (41%), Gaps = 8/73 (10%)
Query 26 AAACAARLSARLPAAAAAAAGPAAAAGQQQQRRLLHTSDLRAAATTISPSEMAKILEQR- 84
AA A R++A P+ GP Q LLH S R +T I E+ ILE +
Sbjct 152 AALMADRIAANFPSTEVYLVGPIGPRSQA----LLHPSVKRTNSTRILKDELHVILEYKQ 207
Query 85 ---ISGWTTQQST 94
+ W S+
Sbjct 208 GEILGDWVAPSSS 220
> CE20196
Length=1413
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 20/65 (30%), Positives = 32/65 (49%), Gaps = 10/65 (15%)
Query 68 AATTISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVGDG-----IARLLGLGNVRAGEL 122
A T S S MAKIL +R+ GW ++ FSV D ++R + + ++ E+
Sbjct 350 AKTLSSASAMAKILYERLFGWIVKRCNDA-----FSVDDTESTCRLSRFIAVLDIAGFEI 404
Query 123 VELQS 127
+E S
Sbjct 405 IEKNS 409
> Hs22044978
Length=258
Score = 30.0 bits (66), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 31/77 (40%), Gaps = 3/77 (3%)
Query 17 PLIR-PLSSAAAACAARLSARLPAAAAAAAGPAAA--AGQQQQRRLLHTSDLRAAATTIS 73
P+IR P S C A L P A P AA +Q L +D + I
Sbjct 64 PVIRFPSGSDTKCCFASLRLYSPNGDVAITSPQAARLHSSKQVTSLKPKTDCKQPTRLIK 123
Query 74 PSEMAKILEQRISGWTT 90
P + K++ +S W+T
Sbjct 124 PKPLMKMVSSYVSLWST 140
> CE01537
Length=394
Score = 27.7 bits (60), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 35/78 (44%), Gaps = 8/78 (10%)
Query 47 PAAAAGQQQQRRLLHTSDLRAAATTISPSEMAKILEQRISGWTTQQSTAGEVGRVFSVG- 105
P + QQ L +RAA +PS++A ++ RIS W Q VG +
Sbjct 290 PTGESAQQ-----LCIDSIRAAVLDTTPSDVAYTVQIRISEWEEQGEVLQIVGEIRCQKP 344
Query 106 -DGIARLLGLGNVRAGEL 122
DG + ++G G R E+
Sbjct 345 RDG-SLIIGKGGKRISEI 361
> Hs18699720
Length=1090
Score = 27.7 bits (60), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 6/61 (9%)
Query 54 QQQRRLLHTSDLRAAATTISPSEM------AKILEQRISGWTTQQSTAGEVGRVFSVGDG 107
QQQ+ LL+ ++ A SE+ KI R++G ++ QS GRV +VG
Sbjct 290 QQQKELLNKRNMEVAMMDKRISELRERLYGKKIQLNRVNGTSSPQSPLSTSGRVAAVGPY 349
Query 108 I 108
I
Sbjct 350 I 350
> CE21619
Length=254
Score = 27.3 bits (59), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 15/30 (50%), Gaps = 0/30 (0%)
Query 38 PAAAAAAAGPAAAAGQQQQRRLLHTSDLRA 67
P AA PA AAG R + TSD+ A
Sbjct 5 PQAATTECSPAMAAGDSSSDRFVGTSDIHA 34
Lambda K H
0.318 0.128 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40