bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
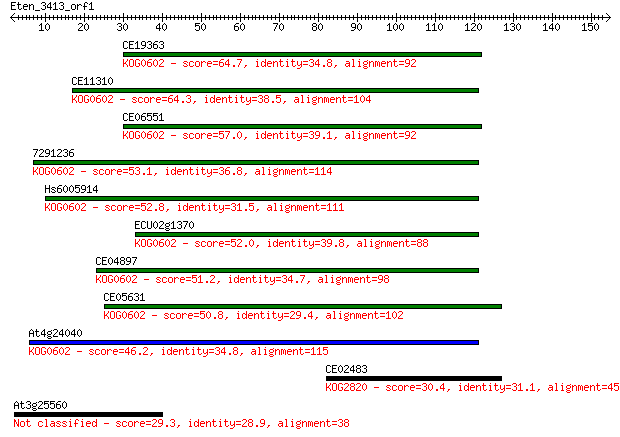
Query= Eten_3413_orf1
Length=154
Score E
Sequences producing significant alignments: (Bits) Value
CE19363 64.7 8e-11
CE11310 64.3 8e-11
CE06551 57.0 1e-08
7291236 53.1 2e-07
Hs6005914 52.8 3e-07
ECU02g1370 52.0 5e-07
CE04897 51.2 7e-07
CE05631 50.8 1e-06
At4g24040 46.2 2e-05
CE02483 30.4 1.4
At3g25560 29.3 3.4
> CE19363
Length=684
Score = 64.7 bits (156), Expect = 8e-11, Method: Composition-based stats.
Identities = 32/93 (34%), Positives = 49/93 (52%), Gaps = 1/93 (1%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQAYNRLGALPEKS 89
+T QQWD PN W P+ M +E + + L + +K++ Q +N + EK
Sbjct 524 ETNQQWDFPNGWSPMNHMIIEGLRKSNNPILQQKAFTLAEKWLETNMQTFNVSDEMWEKY 583
Query 90 NSLIS-GSCGSGGDYKCQIGFGWSIGVALELLF 121
N G +GG+Y+ Q GFGW+ G AL+L+F
Sbjct 584 NVKEPLGKLATGGEYEVQAGFGWTNGAALDLIF 616
> CE11310
Length=570
Score = 64.3 bits (155), Expect = 8e-11, Method: Composition-based stats.
Identities = 40/107 (37%), Positives = 56/107 (52%), Gaps = 8/107 (7%)
Query 17 GLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACT 76
GL S +S+TQ QWDK N WPP++ M +E D L+ + M ++
Sbjct 435 GLPTSLAMSSTQ-----QWDKENAWPPMIHMVIEGFRTTGDIKLMKVAEKMATSWLTGTY 489
Query 77 QAYNRLGALPEKSN---SLISGSCGSGGDYKCQIGFGWSIGVALELL 120
Q++ R A+ EK N S G GG+Y+ Q GFGW+ GV L+LL
Sbjct 490 QSFIRTHAMFEKYNVTPHTEETSGGGGGEYEVQTGFGWTNGVILDLL 536
> CE06551
Length=588
Score = 57.0 bits (136), Expect = 1e-08, Method: Composition-based stats.
Identities = 36/102 (35%), Positives = 51/102 (50%), Gaps = 19/102 (18%)
Query 30 KTGQQWDKPNCWPPLVQMAVEAI-----INLEDQALLSAIK-CMTDKYIFACT----QAY 79
++ QQWD PN W P M +E + ++D+ L A K M + +F T + Y
Sbjct 446 ESDQQWDFPNGWSPNNHMIIEGLRKSANPEMQDKGFLIASKWVMGNFRVFYETGHMWEKY 505
Query 80 NRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELLF 121
N +G+ P+ GSGG+Y Q GFGWS G L+LL
Sbjct 506 NVIGSYPQP---------GSGGEYDVQDGFGWSNGAILDLLL 538
> 7291236
Length=596
Score = 53.1 bits (126), Expect = 2e-07, Method: Composition-based stats.
Identities = 42/117 (35%), Positives = 66/117 (56%), Gaps = 5/117 (4%)
Query 7 SSAQFLVEKSGLWDSW--GLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLED-QALLSA 63
+S +E++ L DS+ G+ T TG+QWD PN W P+ + VE + NL +A +
Sbjct 460 ASVMAYIERNKL-DSFPGGVPNTLSYTGEQWDAPNVWAPMQYILVEGLNNLNTPEAKNMS 518
Query 64 IKCMTDKYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
+K T +++ A+++ + EK N+ G G GG+Y+ Q GFGWS GV +E L
Sbjct 519 LKWAT-RWVKTNFAAFSKDRHMYEKYNADEFGVGGGGGEYEVQTGFGWSNGVIIEWL 574
> Hs6005914
Length=583
Score = 52.8 bits (125), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 53/111 (47%), Gaps = 0/111 (0%)
Query 10 QFLVEKSGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTD 69
++L + L +G+ + KTGQQWD PN W PL + + + + +
Sbjct 437 KYLEDNRILTYQYGIPTSLQKTGQQWDFPNAWAPLQDLVIRGLAKAPLRRAQEVAFQLAQ 496
Query 70 KYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
+I Y++ A+ EK + G G GG+Y+ Q GFGW GV L LL
Sbjct 497 NWIRTNFDVYSQKSAMYEKYDVSNGGQPGGGGEYEVQEGFGWDEGVVLMLL 547
> ECU02g1370
Length=659
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 35/102 (34%), Positives = 48/102 (47%), Gaps = 27/102 (26%)
Query 33 QQWDKPNCWPPLV-------------QMAVE-AIINLEDQALLSAIKCMTDKYIFACTQA 78
+QWD PN WPPLV QMA+ A LE+ ++ ++I +T + IF
Sbjct 497 EQWDYPNVWPPLVHIMTFFLERIGERQMALHLARSLLENISVSTSISDVTRRGIF----- 551
Query 79 YNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELL 120
EK N G G G+Y Q+GFGW+ G A+ L
Sbjct 552 --------EKYNCERVGDSGYKGEYAPQVGFGWTNGSAIHFL 585
> CE04897
Length=585
Score = 51.2 bits (121), Expect = 7e-07, Method: Composition-based stats.
Identities = 34/103 (33%), Positives = 50/103 (48%), Gaps = 11/103 (10%)
Query 23 GLSATQVKTGQQWDKPNCWPPLVQMAVEAI--INLEDQALLSAIKCMTDKYIFACTQAYN 80
G+ + V +G+QWD PN WPP + +E + + E+ AL K + + T
Sbjct 422 GIPVSLVNSGEQWDFPNSWPPTTWVLLEGLRKVGQEELALSLVEKWVQKNFNMWRTSG-- 479
Query 81 RLGALPEKSNSLISGSC---GSGGDYKCQIGFGWSIGVALELL 120
G + EK N + C GG+Y Q GFGW+ GV L+ L
Sbjct 480 --GRMFEKYN--VVSPCFKVKGGGEYVMQEGFGWTNGVILDFL 518
> CE05631
Length=638
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 30/108 (27%), Positives = 53/108 (49%), Gaps = 6/108 (5%)
Query 25 SATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQALLSAIKCMTDKYIFACTQA-YNRLG 83
S+ ++ QQWD PN W P +++ + + L K ++I YN +
Sbjct 460 SSLPAQSTQQWDFPNVWAPNQHFVIQSFMACNNSFLQQEAKKQAMEFIETVYNGMYNPIA 519
Query 84 ALP----EKSNSL-ISGSCGSGGDYKCQIGFGWSIGVALELLFAKRGT 126
L EK ++ +G+ G+GG+Y Q GFGW+ G ++L++ R +
Sbjct 520 GLDGGVWEKYDARSTNGAPGAGGEYVVQEGFGWTNGAVMDLIWTLRDS 567
> At4g24040
Length=626
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 40/122 (32%), Positives = 57/122 (46%), Gaps = 7/122 (5%)
Query 6 ISSAQFLVEK-------SGLWDSWGLSATQVKTGQQWDKPNCWPPLVQMAVEAIINLEDQ 58
I+S + LV+K SGL G+ + +GQQWD PN W P +M V + +
Sbjct 492 INSDENLVKKVVTALKNSGLIAPAGILTSLTNSGQQWDSPNGWAPQQEMIVTGLGRSSVK 551
Query 59 ALLSAIKCMTDKYIFACTQAYNRLGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALE 118
+ + ++I + Y + G + EK G G GG+Y Q GFGWS GV L
Sbjct 552 EAKEMAEDIARRWIKSNYLVYKKSGTIHEKLKVTELGEYGGGGEYMPQTGFGWSNGVILA 611
Query 119 LL 120
L
Sbjct 612 FL 613
> CE02483
Length=703
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 25/45 (55%), Gaps = 3/45 (6%)
Query 82 LGALPEKSNSLISGSCGSGGDYKCQIGFGWSIGVALELLFAKRGT 126
+G +P K+ +++ G CGSG +K G G ++ E+ K+ T
Sbjct 646 IGTIPTKNPNILVGGCGSGSGFKVAPGIGKALA---EMAAGKKTT 687
> At3g25560
Length=531
Score = 29.3 bits (64), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 11/38 (28%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 2 HKRQISSAQFLVEKSGLWDSWGLSATQVKTGQQWDKPN 39
H+ ++S ++E GL + W S+ + +T + + KPN
Sbjct 466 HRPKMSEVVRMLEGDGLVEKWEASSQRAETNRSYSKPN 503
Lambda K H
0.320 0.132 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1997677330
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40