bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3442_orf2
Length=226
Score E
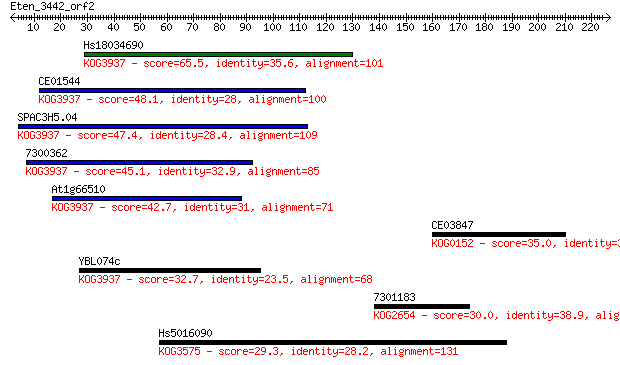
Sequences producing significant alignments: (Bits) Value
Hs18034690 65.5 9e-11
CE01544 48.1 2e-05
SPAC3H5.04 47.4 2e-05
7300362 45.1 1e-04
At1g66510 42.7 6e-04
CE03847 35.0 0.13
YBL074c 32.7 0.57
7301183 30.0 4.5
Hs5016090 29.3 6.3
> Hs18034690
Length=384
Score = 65.5 bits (158), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 54/101 (53%), Gaps = 4/101 (3%)
Query 29 VLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPELFSKFIDTFYAQLEQAP 88
VL ELQ AF+ +LG+ Y + HWK LL L+C SE A+ + L+ I Y QL + P
Sbjct 251 VLGELQFAFVCFLLGNVYEAFEHWKRLLNLLCRSEAAMMKHHTLYINLISILYHQLGEIP 310
Query 89 DELTELEPLQQNNLLTSCCAALLEFCCSVDRQAARAALKQQ 129
+ ++ + Q+N LTS CS+ A A L+++
Sbjct 311 ADFF-VDIVSQDNFLTSTLQVFFSSACSI---AVDATLRKK 347
> CE01544
Length=357
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 51/100 (51%), Gaps = 1/100 (1%)
Query 12 SDALLLLIREHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPE 71
SD L L+R + +LAE+Q+AF+ + G + WK ++ L+ +L E
Sbjct 224 SDRLYRLLRALGGDWKQLLAEMQIAFVCFLQGQVFEGFEQWKRIIHLMSCCPNSLGSEKE 283
Query 72 LFSKFIDTFYAQLEQAPDELTELEPLQQNNLLTSCCAALL 111
LF FI + QL++ P + ++ + ++N LT+ + L
Sbjct 284 LFMSFIRVLFFQLKECPTDFF-VDIVSRDNFLTTTLSMLF 322
> SPAC3H5.04
Length=346
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 31/110 (28%), Positives = 54/110 (49%), Gaps = 1/110 (0%)
Query 4 RRPQCTDRSDALLLLIRE-HQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNS 62
R Q D+S L++ + + LAEL ++F++ + HY +L HWK++L L+ S
Sbjct 168 RSKQAIDKSFLFQRLVQSVWNDNPISALAELSISFLSYSILSHYGALEHWKNMLSLLLQS 227
Query 63 ERALYLYPELFSKFIDTFYAQLEQAPDELTELEPLQQNNLLTSCCAALLE 112
PE ++ F++ F QL + E + + +L SC +L E
Sbjct 228 YELAETEPEFYASFLELFKLQLSSLSESDLETSAIFEKGVLLSCLDSLSE 277
> 7300362
Length=379
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 45/88 (51%), Gaps = 10/88 (11%)
Query 7 QCTDRSDALLLLIREHQEGFA---AVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSE 63
C D D LL EGF ++ ELQ+A+ ++G+ SL HW+ LL L+ +S+
Sbjct 235 DCIDAVDKLL-------EGFETADGLIEELQLAYAFFLVGYSVESLAHWRKLLGLLAHSQ 287
Query 64 RALYLYPELFSKFIDTFYAQLEQAPDEL 91
A+ + + K+ + QL P+EL
Sbjct 288 TAVTKHKLTYMKYSEVLAHQLPHLPEEL 315
> At1g66510
Length=399
Score = 42.7 bits (99), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 1/72 (1%)
Query 17 LLIREHQEGFAAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALY-LYPELFSK 75
+L +E+++ +L ELQ +F+A ++G S + WK ++ L+ A + +LF+K
Sbjct 225 VLSKEYKDSEDLLLGELQFSFVAFLMGQSLESFMQWKSIVSLLLGCTSAPFQTRSQLFTK 284
Query 76 FIDTFYAQLEQA 87
FI Y QL+
Sbjct 285 FIKVIYHQLKYG 296
> CE03847
Length=724
Score = 35.0 bits (79), Expect = 0.13, Method: Composition-based stats.
Identities = 19/50 (38%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query 160 PDAMQTEQTQQQPPAAAEATAPAAEAAAAAQAAAAEAEIDRAVAARLAAL 209
PD + + +Q+P A A AT AAA Q AE+++D+A+ A LA++
Sbjct 154 PDGEEITKGEQKPAAKA-ATVDTVALAAAVQQKKAESDLDKAMKATLASM 202
> YBL074c
Length=355
Score = 32.7 bits (73), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 16/69 (23%), Positives = 36/69 (52%), Gaps = 5/69 (7%)
Query 27 AAVLAELQMAFIAGVLGHHYPSLVHWKDLLQLVCNSERALYLYPE-LFSKFIDTFYAQLE 85
+ ELQ AF+ + +Y S + W +++L+C+S P+ + K + Y Q++
Sbjct 218 SNYFGELQFAFLNAMFFGNYGSSLQWHAMIELICSSATV----PKHMLDKLDEILYYQIK 273
Query 86 QAPDELTEL 94
P++ +++
Sbjct 274 TLPEQYSDI 282
> 7301183
Length=647
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 138 TPAAAAAGKAAPTRRREVHDRHPDAMQTEQTQQQPP 173
+P GKA+PTRRR D+ P + + Q PP
Sbjct 231 SPPRKRTGKASPTRRRRDSDQSPPRRRRSNSDQSPP 266
> Hs5016090
Length=470
Score = 29.3 bits (64), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 37/132 (28%), Positives = 52/132 (39%), Gaps = 24/132 (18%)
Query 57 QLVCNSERALYLYPELFSKFIDTFYAQLEQAPDELTELEPL-QQNNLLTSCCAALLEFCC 115
Q+VC + A L F K++ F ++ PL QQ L+ +C A+LL
Sbjct 252 QVVCEAASAGLLKTLRFVKYLPCF------------QVLPLDQQLVLVRNCWASLLMLEL 299
Query 116 SVDRQAARAALKQQQVEAAAAATPAAAAAGKAAPTRRREVHDRHPDAMQTEQTQQQPPAA 175
+ DR Q E + P+ K TRRRE P + T Q PPA
Sbjct 300 AQDRL---------QFETVEVSEPSMLQ--KILTTRRRETGGNEPLPVPTLQHHLAPPAE 348
Query 176 AEATAPAAEAAA 187
A A++ A
Sbjct 349 ARKVPSASQVQA 360
Lambda K H
0.318 0.127 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4312910206
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40