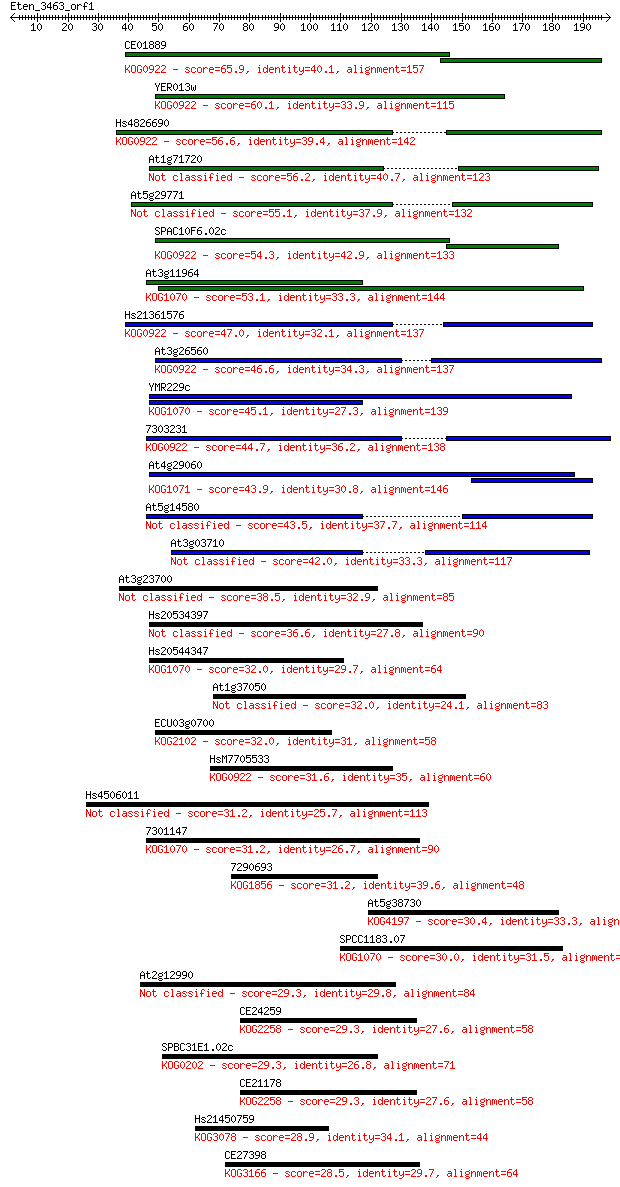
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3463_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
CE01889 65.9 5e-11
YER013w 60.1 3e-09
Hs4826690 56.6 3e-08
At1g71720 56.2 4e-08
At5g29771 55.1 1e-07
SPAC10F6.02c 54.3 2e-07
At3g11964 53.1 4e-07
Hs21361576 47.0 2e-05
At3g26560 46.6 4e-05
YMR229c 45.1 1e-04
7303231 44.7 1e-04
At4g29060 43.9 2e-04
At5g14580 43.5 3e-04
At3g03710 42.0 9e-04
At3g23700 38.5 0.008
Hs20534397 36.6 0.038
Hs20544347 32.0 0.81
At1g37050 32.0 0.82
ECU03g0700 32.0 0.86
HsM7705533 31.6 1.2
Hs4506011 31.2 1.3
7301147 31.2 1.5
7290693 31.2 1.6
At5g38730 30.4 2.1
SPCC1183.07 30.0 2.8
At2g12990 29.3 4.7
CE24259 29.3 5.6
SPBC31E1.02c 29.3 5.8
CE21178 29.3 5.8
Hs21450759 28.9 6.8
CE27398 28.5 8.2
> CE01889
Length=1200
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 40/109 (36%), Positives = 61/109 (55%), Gaps = 3/109 (2%)
Query 39 RPQATSGVGAVLKGVVKNIRPFGAFILTSQY--ARECLLHISNISRQRVENVEDALKMGQ 96
R + +G + G V +I+ FGAFI + +E L+HIS I +RV+ V D LK G+
Sbjct 226 RKSEVAEIGKIYDGRVNSIQSFGAFITLEGFRQKQEGLVHISQIRNERVQTVADVLKRGE 285
Query 97 EVWVKVLKEEPDGRVSVSMKDVNQEDGSEITSLFDSFNNRGVGGGVKIP 145
V VKV K E +G++S+SMK+V+Q G ++ N +G + P
Sbjct 286 NVKVKVNKIE-NGKISLSMKEVDQNSGEDLNPRETDLNPDAIGVRPRTP 333
Score = 48.1 bits (113), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/55 (41%), Positives = 36/55 (65%), Gaps = 2/55 (3%)
Query 143 KIPELNSIHRGTVKRIQAFGAFVALDGF--DRDGLLHISCISKDKIDKVDDVAHR 195
++ E+ I+ G V IQ+FGAF+ L+GF ++GL+HIS I +++ V DV R
Sbjct 229 EVAEIGKIYDGRVNSIQSFGAFITLEGFRQKQEGLVHISQIRNERVQTVADVLKR 283
> YER013w
Length=1145
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 39/128 (30%), Positives = 69/128 (53%), Gaps = 13/128 (10%)
Query 49 VLKGVVKNIRPFGAFI-LTSQYAREC--LLHISNISRQRVENVEDALKMGQEVWVKVLKE 105
V +G V+NI FG F+ + + C L+HIS +S QR + D ++ GQ ++V+V+K
Sbjct 180 VYEGKVRNITTFGCFVQIFGTRMKNCDGLVHISEMSDQRTLDPHDVVRQGQHIFVEVIKI 239
Query 106 EPDGRVSVSMKDVNQEDGSEITSLFDSFNNRG----------VGGGVKIPELNSIHRGTV 155
+ +G++S+SMK+++Q G +S +RG + +K L S R +
Sbjct 240 QNNGKISLSMKNIDQHSGEIRKRNTESVEDRGRSNDAHTSRNMKNKIKRRALTSPERWEI 299
Query 156 KRIQAFGA 163
+++ A GA
Sbjct 300 RQLIASGA 307
> Hs4826690
Length=1220
Score = 56.6 bits (135), Expect = 3e-08, Method: Composition-based stats.
Identities = 37/94 (39%), Positives = 52/94 (55%), Gaps = 8/94 (8%)
Query 36 PPKRPQATSGVGAVLKGVVKNIRPFGAFILTSQYAR--ECLLHISNISRQ-RVENVEDAL 92
PP+ P +G + G V +I FG F+ + E L+HIS + R+ RV NV D +
Sbjct 258 PPEEPT----IGDIYNGKVTSIMQFGCFVQLEGLRKRWEGLVHISELRREGRVANVADVV 313
Query 93 KMGQEVWVKVLKEEPDGRVSVSMKDVNQEDGSEI 126
GQ V VKVL + S+SMKDV+QE G ++
Sbjct 314 SKGQRVKVKVLSFT-GTKTSLSMKDVDQETGEDL 346
Score = 35.8 bits (81), Expect = 0.065, Method: Composition-based stats.
Identities = 19/54 (35%), Positives = 31/54 (57%), Gaps = 3/54 (5%)
Query 145 PELNSIHRGTVKRIQAFGAFVALDGFDR--DGLLHISCISKD-KIDKVDDVAHR 195
P + I+ G V I FG FV L+G + +GL+HIS + ++ ++ V DV +
Sbjct 262 PTIGDIYNGKVTSIMQFGCFVQLEGLRKRWEGLVHISELRREGRVANVADVVSK 315
> At1g71720
Length=487
Score = 56.2 bits (134), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 48/77 (62%), Gaps = 0/77 (0%)
Query 47 GAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEE 106
G +L+G V I P+GA + +R LLHISNI+R+R+ +V D L++ + V V V+K
Sbjct 349 GTLLEGTVVKILPYGAQVKLGDSSRSGLLHISNITRRRIGSVSDVLQVDESVKVLVVKSL 408
Query 107 PDGRVSVSMKDVNQEDG 123
++S+S+ D+ E G
Sbjct 409 FPDKISLSIADLESEPG 425
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 27/46 (58%), Gaps = 0/46 (0%)
Query 149 SIHRGTVKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDDVAH 194
++ GTV +I +GA V L R GLLHIS I++ +I V DV
Sbjct 350 TLLEGTVVKILPYGAQVKLGDSSRSGLLHISNITRRRIGSVSDVLQ 395
> At5g29771
Length=416
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/87 (39%), Positives = 51/87 (58%), Gaps = 3/87 (3%)
Query 41 QATSGVGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWV 100
QA G+G+V+ GVV++++P+GAFI LLH+S IS RV ++ L+ G + V
Sbjct 258 QAQLGIGSVVLGVVQSLKPYGAFIDIGGI--NGLLHVSQISHDRVSDIATVLQPGDTLKV 315
Query 101 KVLKEEPD-GRVSVSMKDVNQEDGSEI 126
+L + D GRVS+S K + G I
Sbjct 316 MILSHDRDRGRVSLSTKKLEPTPGDMI 342
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 29/46 (63%), Gaps = 2/46 (4%)
Query 147 LNSIHRGTVKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDDV 192
+ S+ G V+ ++ +GAF+ + G + GLLH+S IS D++ + V
Sbjct 263 IGSVVLGVVQSLKPYGAFIDIGGIN--GLLHVSQISHDRVSDIATV 306
> SPAC10F6.02c
Length=1168
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 36/101 (35%), Positives = 58/101 (57%), Gaps = 4/101 (3%)
Query 49 VLKGVVKNIRPFGAFILTSQYAREC--LLHISNISRQ-RVENVEDALKMGQEVWVKVLK- 104
+ GVV I+ FGAF+ + + L+HISNI R+++ +A+ GQ V+VKV++
Sbjct 209 IYSGVVSGIKDFGAFVTLDGFRKRTDGLVHISNIQLNGRLDHPSEAVSYGQPVFVKVIRI 268
Query 105 EEPDGRVSVSMKDVNQEDGSEITSLFDSFNNRGVGGGVKIP 145
+E R+S+SMK+VNQ G ++ S + + G IP
Sbjct 269 DESAKRISLSMKEVNQVTGEDLNPDQVSRSTKKGSGANAIP 309
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/39 (56%), Positives = 26/39 (66%), Gaps = 2/39 (5%)
Query 145 PELNSIHRGTVKRIQAFGAFVALDGFDR--DGLLHISCI 181
P L I+ G V I+ FGAFV LDGF + DGL+HIS I
Sbjct 204 PTLYGIYSGVVSGIKDFGAFVTLDGFRKRTDGLVHISNI 242
> At3g11964
Length=1838
Score = 53.1 bits (126), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 1/72 (1%)
Query 46 VGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLK- 104
VG ++ G ++ + PFG FI Q L HIS +S R+ENV+ K G+ V K+LK
Sbjct 1376 VGDMISGRIRRVEPFGLFIDIDQTGMVGLCHISQLSDDRMENVQARYKAGESVRAKILKL 1435
Query 105 EEPDGRVSVSMK 116
+E R+S+ MK
Sbjct 1436 DEEKKRISLGMK 1447
Score = 51.6 bits (122), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 70/141 (49%), Gaps = 13/141 (9%)
Query 50 LKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEP-D 108
++G VKN G FI+ S+ E + +SN+ V+ E +G+ V +VL EP
Sbjct 1290 VQGYVKNTMSKGCFIILSRTV-EAKVRLSNLCDTFVKEPEKEFPVGKLVTGRVLNVEPLS 1348
Query 109 GRVSVSMKDVNQEDGSEITSLFDSFNNRGVGGGVKIPELNSIHRGTVKRIQAFGAFVALD 168
R+ V++K VN G + +D +K + + G ++R++ FG F+ +D
Sbjct 1349 KRIEVTLKTVNA-GGRPKSESYD----------LKKLHVGDMISGRIRRVEPFGLFIDID 1397
Query 169 GFDRDGLLHISCISKDKIDKV 189
GL HIS +S D+++ V
Sbjct 1398 QTGMVGLCHISQLSDDRMENV 1418
> Hs21361576
Length=241
Score = 47.0 bits (110), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 52/94 (55%), Gaps = 6/94 (6%)
Query 39 RPQATSGVGA---VLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMG 95
RP+ + A + +G V + +GAFI ++ L+H +++S RV+ + + +G
Sbjct 5 RPETMENLPALYTIFQGEVAMVTDYGAFIKIPGCRKQGLVHRTHMSSCRVDKPSEIVDVG 64
Query 96 QEVWVKVLKEEPDG---RVSVSMKDVNQEDGSEI 126
+VWVK++ E +VS+SMK VNQ G ++
Sbjct 65 DKVWVKLIGREMKNDRIKVSLSMKVVNQGTGKDL 98
Score = 36.6 bits (83), Expect = 0.032, Method: Compositional matrix adjust.
Identities = 15/49 (30%), Positives = 30/49 (61%), Gaps = 0/49 (0%)
Query 144 IPELNSIHRGTVKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDDV 192
+P L +I +G V + +GAF+ + G + GL+H + +S ++DK ++
Sbjct 12 LPALYTIFQGEVAMVTDYGAFIKIPGCRKQGLVHRTHMSSCRVDKPSEI 60
> At3g26560
Length=1168
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 49/82 (59%), Gaps = 2/82 (2%)
Query 49 VLKGVVKNIRPFGAFILTSQY-ARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEP 107
V KG V + G F+ ++ +E L+H+S ++ +RV+ ++ +K EV+VKV+
Sbjct 216 VYKGRVTRVMDAGCFVQFDKFRGKEGLVHVSQMATRRVDKAKEFVKRDMEVYVKVISISS 275
Query 108 DGRVSVSMKDVNQEDGSEITSL 129
D + S+SM+DV+Q G ++ L
Sbjct 276 D-KYSLSMRDVDQNTGRDLIPL 296
Score = 44.3 bits (103), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 140 GGVKIPELNSIHRGTVKRIQAFGAFVALDGFD-RDGLLHISCISKDKIDKVDDVAHR 195
GG PEL +++G V R+ G FV D F ++GL+H+S ++ ++DK + R
Sbjct 206 GGANEPELYQVYKGRVTRVMDAGCFVQFDKFRGKEGLVHVSQMATRRVDKAKEFVKR 262
> YMR229c
Length=1729
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 38/143 (26%), Positives = 69/143 (48%), Gaps = 20/143 (13%)
Query 47 GAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEE 106
G ++ G+VKN+ G F+ S+ E + +S +S ++ + K Q V KV+ +
Sbjct 1177 GEIVDGIVKNVNDKGIFVYLSRKV-EAFVPVSKLSDSYLKEWKKFYKPMQYVLGKVVTCD 1235
Query 107 PDGRVSVSMKDVNQEDGSEIT---SLFDSFNNRGVGGGVKIPELNSIHRGTVKRIQAFGA 163
D R+S+++++ SEI + ++++ G + GT+K + FG
Sbjct 1236 EDSRISLTLRE------SEINGDLKVLKTYSDIKAG---------DVFEGTIKSVTDFGV 1280
Query 164 FVALDG-FDRDGLLHISCISKDK 185
FV LD + GL HI+ I+ K
Sbjct 1281 FVKLDNTVNVTGLAHITEIADKK 1303
Score = 35.0 bits (79), Expect = 0.087, Method: Compositional matrix adjust.
Identities = 22/72 (30%), Positives = 40/72 (55%), Gaps = 2/72 (2%)
Query 47 GAVLKGVVKNIRPFGAFI-LTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKE 105
G V +G +K++ FG F+ L + L HI+ I+ ++ E++ +G V VLK
Sbjct 1265 GDVFEGTIKSVTDFGVFVKLDNTVNVTGLAHITEIADKKPEDLSALFGVGDRVKAIVLKT 1324
Query 106 EPDGR-VSVSMK 116
P+ + +S+S+K
Sbjct 1325 NPEKKQISLSLK 1336
> 7303231
Length=1242
Score = 44.7 bits (104), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 50/89 (56%), Gaps = 8/89 (8%)
Query 46 VGAVLKGVVKNIRPFGAFI----LTSQYARECLLHISNISRQ-RVENVEDALKMGQEVWV 100
G + G + NI PFG F+ L ++ E L+HIS + + RV +V + + Q V V
Sbjct 284 AGKIYSGKIANIVPFGCFVQLFGLRKRW--EGLVHISQLRAEGRVTDVTEVVTRNQTVKV 341
Query 101 KVLKEEPDGRVSVSMKDVNQEDGSEITSL 129
KV+ +VS+SMK+V+Q+ G ++ L
Sbjct 342 KVMSIT-GQKVSLSMKEVDQDSGKDLNPL 369
Score = 32.3 bits (72), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 3/57 (5%)
Query 145 PELNSIHRGTVKRIQAFGAFVALDGFDR--DGLLHISCISKD-KIDKVDDVAHRKHV 198
PE I+ G + I FG FV L G + +GL+HIS + + ++ V +V R
Sbjct 282 PEAGKIYSGKIANIVPFGCFVQLFGLRKRWEGLVHISQLRAEGRVTDVTEVVTRNQT 338
> At4g29060
Length=953
Score = 43.9 bits (102), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 43/152 (28%), Positives = 76/152 (50%), Gaps = 14/152 (9%)
Query 47 GAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEE 106
GA G V+ I+PFGAF+ + + L+H+S +S V++V + +GQEV V++++ +
Sbjct 136 GATFTGKVRAIQPFGAFVDFGAFT-DGLVHVSQLSDNFVKDVSSVVTIGQEVKVRLVEAD 194
Query 107 PDG-RVSVSMKDVN-----QEDGSEITSLFDSFNNRGVGGGVKIPELNS------IHRGT 154
+ R+S++M++ + Q GS+ + GG K NS + G
Sbjct 195 IESKRISLTMRENDDPPKRQSGGSDKPRSGGKRDGSKGGGQRKGEGFNSKFAKGQMLDGV 254
Query 155 VKRIQAFGAFVALDGFDRDGLLHISCISKDKI 186
VK + GAF+ + G +G L + + D I
Sbjct 255 VKNLTRSGAFITI-GEGEEGFLPTAEEADDGI 285
Score = 32.7 bits (73), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Query 153 GTVKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDDV 192
G V+ IQ FGAFV F DGL+H+S +S + + V V
Sbjct 141 GKVRAIQPFGAFVDFGAFT-DGLVHVSQLSDNFVKDVSSV 179
> At5g14580
Length=991
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 40/71 (56%), Gaps = 1/71 (1%)
Query 46 VGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKE 105
VG V KG V +I+ +GAF+ ++ LLH+S +S + V V D L +GQ + ++
Sbjct 677 VGGVYKGTVSSIKEYGAFV-EFPGGQQGLLHMSELSHEPVSKVSDVLDIGQCITTMCIET 735
Query 106 EPDGRVSVSMK 116
+ G + +S K
Sbjct 736 DVRGNIKLSRK 746
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 29/43 (67%), Gaps = 1/43 (2%)
Query 150 IHRGTVKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDDV 192
+++GTV I+ +GAFV G + GLLH+S +S + + KV DV
Sbjct 680 VYKGTVSSIKEYGAFVEFPG-GQQGLLHMSELSHEPVSKVSDV 721
> At3g03710
Length=948
Score = 42.0 bits (97), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 40/63 (63%), Gaps = 1/63 (1%)
Query 54 VKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSV 113
+K++ P+GAF+ + RE L HIS +S + + EDA K+G + VK+++ G++ +
Sbjct 796 IKSMAPYGAFVEIAP-GREGLCHISELSAEWLAKPEDAYKVGDRIDVKLIEVNEKGQLRL 854
Query 114 SMK 116
S++
Sbjct 855 SVR 857
Score = 32.0 bits (71), Expect = 0.90, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 31/55 (56%), Gaps = 2/55 (3%)
Query 138 VGGGVKIPELNSIHRGT-VKRIQAFGAFVALDGFDRDGLLHISCISKDKIDKVDD 191
+ G +P + I+R +K + +GAFV + R+GL HIS +S + + K +D
Sbjct 778 ISGLTMVPSVGDIYRNCEIKSMAPYGAFVEI-APGREGLCHISELSAEWLAKPED 831
> At3g23700
Length=371
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 47/86 (54%), Gaps = 6/86 (6%)
Query 37 PKRPQATSGVGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQ 96
PK Q + VG V G V ++ +GAFI + R +H+S +S V++V D L+ G
Sbjct 178 PKYSQNVN-VGDVFNGRVGSVEDYGAFI----HLRFDDVHVSEVSWDYVQDVRDVLRDGD 232
Query 97 EVWVKVLK-EEPDGRVSVSMKDVNQE 121
EV V V ++ R+++S+K + +
Sbjct 233 EVRVIVTNIDKEKSRITLSIKQLEDD 258
> Hs20534397
Length=783
Score = 36.6 bits (83), Expect = 0.038, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 46/92 (50%), Gaps = 6/92 (6%)
Query 47 GAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVEN-VEDALKMGQEVWVKVLKE 105
GAV + IR G + LLH + + ++++++ L++GQE+ VK
Sbjct 679 GAVYTATITEIRDTGVMVKLYPNMTAVLLHNTQLDQRKIKHPTALGLEVGQEIQVKYFGR 738
Query 106 EP-DGRVSVSMKDVNQEDGSEITSLFDSFNNR 136
+P DGR+ +S K + S T++ + N+R
Sbjct 739 DPADGRMRLSRKVLQ----SPATTVVRTLNDR 766
> Hs20544347
Length=1871
Score = 32.0 bits (71), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 19/66 (28%), Positives = 36/66 (54%), Gaps = 5/66 (7%)
Query 47 GAVLKGVVKNIRPFGAFILTSQYARECL--LHISNISRQRVENVEDALKMGQEVWVKVLK 104
GAV+KG V I+ +G + + R + +H+++I ++N E +G EV +VL
Sbjct 453 GAVVKGTVLTIKSYGMLVKVGEQMRGLVPPMHLADI---LMKNPEKKYHIGDEVKCRVLL 509
Query 105 EEPDGR 110
+P+ +
Sbjct 510 CDPEAK 515
> At1g37050
Length=620
Score = 32.0 bits (71), Expect = 0.82, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 42/83 (50%), Gaps = 3/83 (3%)
Query 68 QYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSVSMKDVNQEDGSEIT 127
+ CL SN+S +V ++ +A+ ++V V+ + EE R+++ + NQ++ +E
Sbjct 244 HFQNTCLALDSNLSHSKVSDMNEAI---EKVMVEKILEEVQARINLPIGGNNQDNATEEE 300
Query 128 SLFDSFNNRGVGGGVKIPELNSI 150
S + R G + IP+ +
Sbjct 301 WSTPSKSTRTPGKTIDIPKFGEV 323
> ECU03g0700
Length=835
Score = 32.0 bits (71), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 11/58 (18%)
Query 49 VLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEE 106
V++G V NI+P G + +Y S + V E +M QEVWVK+ +++
Sbjct 769 VVRGFVTNIKPNGIVVYVPEY-----------SLEEVVVSEGCFEMFQEVWVKIRRDD 815
> HsM7705533
Length=212
Score = 31.6 bits (70), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 21/63 (33%), Positives = 34/63 (53%), Gaps = 3/63 (4%)
Query 67 SQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDG---RVSVSMKDVNQEDG 123
SQ + I +S RV+ + + +G +VWVK++ E +VS+SMK VNQ G
Sbjct 7 SQAVGSKVWSIELMSSCRVDKPSEIVDVGDKVWVKLIGREMKNDRIKVSLSMKVVNQGTG 66
Query 124 SEI 126
++
Sbjct 67 KDL 69
> Hs4506011
Length=1122
Score = 31.2 bits (69), Expect = 1.3, Method: Composition-based stats.
Identities = 29/121 (23%), Positives = 51/121 (42%), Gaps = 11/121 (9%)
Query 26 EGDAFSRSPSPPKRPQATSGVGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRV 85
E + P P +P+ S ++ + R G T +Y+ E L + + +
Sbjct 959 EIQGIEKHPYPESKPEEVSRSSGIVTSGSRKERCIGQIFQTEEYSVEKSLGPMILINKPL 1018
Query 86 ENVEDA-------LKMGQEVWVKVLKEEPDGRVSVSMK-DVNQEDGSEITSLFDSFNNRG 137
EN+E+A + GQ ++ KE D S S+ + +Q G+E SLF + N
Sbjct 1019 ENMEEARHENEGLVSSGQSLYTSGEKES-DSSASTSLPVEESQAQGNE--SLFSKYTNSK 1075
Query 138 V 138
+
Sbjct 1076 I 1076
> 7301147
Length=1220
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 5/95 (5%)
Query 46 VGAVLKGVVKNI--RPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVL 103
VG +++ VK+ + + Y + +H+ + V+N L Q +WVKVL
Sbjct 698 VGDIIRSYVKHATDQVVDLMVCVRNYNKPVKVHVKMLRLNAVKNAPVELVPEQLLWVKVL 757
Query 104 KEEPDGR---VSVSMKDVNQEDGSEITSLFDSFNN 135
+E + + VS + DV D S+ L + + N
Sbjct 758 SKEVETKTLTVSAKLTDVWSGDLSDTAKLVEGYLN 792
> 7290693
Length=1831
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 33/55 (60%), Gaps = 8/55 (14%)
Query 74 LLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSVS-------MKDVNQE 121
+HI N+S ++V N E+ +++ Q + V+++K + D R SV +KDVN E
Sbjct 1243 FIHIKNLSDRQVRNPEERVRVSQMIHVRIIKIDID-RFSVECSSRTADLKDVNNE 1296
> At5g38730
Length=596
Score = 30.4 bits (67), Expect = 2.1, Method: Composition-based stats.
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 10/67 (14%)
Query 119 NQEDGSEITSLFDSFNNRGVGGGVKIPELNSIHRGTVKRI----QAFGAFVALDGFDRDG 174
NQ EIT L + F RG+ V +++RG ++RI Q A V + ++ G
Sbjct 459 NQNKQDEITKLLEEFEKRGLCADV------ALYRGLIRRICKLEQVDYAKVLFESMEKKG 512
Query 175 LLHISCI 181
L+ S I
Sbjct 513 LVGDSVI 519
> SPCC1183.07
Length=1690
Score = 30.0 bits (66), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 13/74 (17%)
Query 110 RVSVSMKDVNQEDGSEITSLFDSFNNRGVGGGVKIPELNSIHRGTVKRIQAFGAFVALDG 169
R+ +S+K +D SEIT +F + VG + GTV ++ +G + +DG
Sbjct 1210 RIEMSLKQSKIKDSSEITK---TFADIAVGSNLD---------GTVVKVGDYGVLIRIDG 1257
Query 170 FDRD-GLLHISCIS 182
D GL H S I+
Sbjct 1258 TDNIVGLCHKSEIA 1271
> At2g12990
Length=209
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 25/86 (29%), Positives = 35/86 (40%), Gaps = 14/86 (16%)
Query 44 SGVGAVLKGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVL 103
+ +G V K F FIL SQ + L + QR EVW+ V+
Sbjct 82 NNLGVVDVNANKTYAKFEPFILASQAGQVSFLSYPRVRSQR------------EVWLSVI 129
Query 104 KEEPDGRVSVSMKDV--NQEDGSEIT 127
K P GR+ + DV QE E++
Sbjct 130 KVNPRGRIVGLVDDVVMQQESIREVS 155
> CE24259
Length=298
Score = 29.3 bits (64), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 30/58 (51%), Gaps = 9/58 (15%)
Query 77 ISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSVSMKDVNQEDGSEITSLFDSFN 134
++ +SR+RV ++ED +K E + R+ +KD + E +I +LF +N
Sbjct 152 LATVSRERVPDMEDVVKWAV---------ENNTRMLFDVKDSDNELVDQIANLFQKYN 200
> SPBC31E1.02c
Length=899
Score = 29.3 bits (64), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/71 (26%), Positives = 38/71 (53%), Gaps = 6/71 (8%)
Query 51 KGVVKNIRPFGAFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDGR 110
KG+ NI+ F F L++ A L+ IS+ V ++ L Q +W+ +L + P +
Sbjct 676 KGIFNNIKNFITFQLSTSVAALSLIAISS-----VFGFQNPLNAMQILWINILMDGPPAQ 730
Query 111 VSVSMKDVNQE 121
S+ ++ V+++
Sbjct 731 -SLGVESVDED 740
> CE21178
Length=356
Score = 29.3 bits (64), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 16/58 (27%), Positives = 30/58 (51%), Gaps = 9/58 (15%)
Query 77 ISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSVSMKDVNQEDGSEITSLFDSFN 134
++ +SR+RV ++ED +K E + R+ +KD + E +I +LF +N
Sbjct 152 LATVSRERVPDMEDVVKWAV---------ENNTRMLFDVKDSDNELVDQIANLFQKYN 200
> Hs21450759
Length=421
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 0/44 (0%)
Query 62 AFILTSQYARECLLHISNISRQRVENVEDALKMGQEVWVKVLKE 105
AFI Q E L H+ +ENVE+ +K+ +E ++ L+E
Sbjct 211 AFIAEMQMVAEILHHLVQRPEDYLENVENIVKLYKETILQTLEE 254
> CE27398
Length=265
Score = 28.5 bits (62), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 72 ECLLHISNISRQRVENVEDALKMGQEVWVKVLKEEPDGRVSVSMKDVNQEDGSEITSLFD 131
E +LH+ + R+ NV A+ G+ V++ + +V++ DVN ED S + L +
Sbjct 166 EIVLHLPALCRK--YNVPYAIIKGKASLGTVVRRKTTA--AVALVDVNPEDKSALNKLVE 221
Query 132 SFNN 135
+ NN
Sbjct 222 TVNN 225
Lambda K H
0.320 0.139 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3407623970
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40