bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3479_orf2
Length=175
Score E
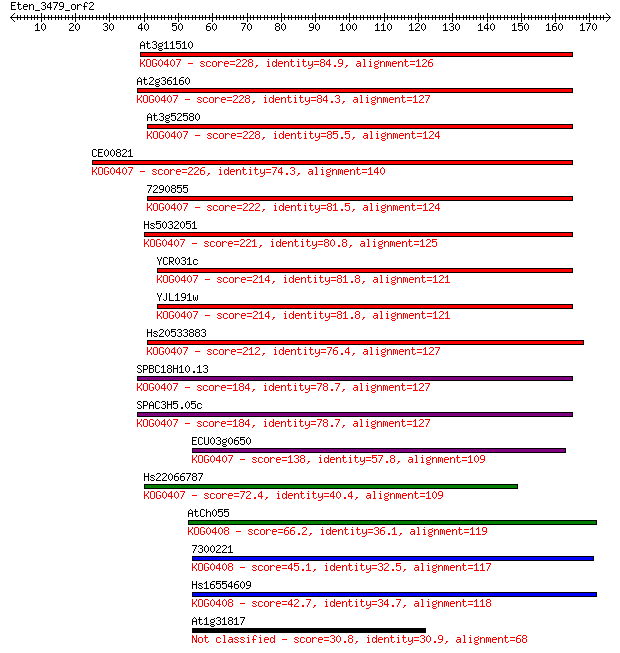
Sequences producing significant alignments: (Bits) Value
At3g11510 228 3e-60
At2g36160 228 4e-60
At3g52580 228 5e-60
CE00821 226 2e-59
7290855 222 3e-58
Hs5032051 221 7e-58
YCR031c 214 6e-56
YJL191w 214 7e-56
Hs20533883 212 3e-55
SPBC18H10.13 184 1e-46
SPAC3H5.05c 184 1e-46
ECU03g0650 138 5e-33
Hs22066787 72.4 5e-13
AtCh055 66.2 3e-11
7300221 45.1 8e-05
Hs16554609 42.7 4e-04
At1g31817 30.8 1.6
> At3g11510
Length=150
Score = 228 bits (582), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 107/126 (84%), Positives = 116/126 (92%), Gaps = 0/126 (0%)
Query 39 AALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAA 98
LGP RE E VFGV HI+ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAA
Sbjct 14 VTLGPSVREGEQVFGVVHIFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAA 73
Query 99 MMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTP 158
M+AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVTP
Sbjct 74 MLAAQDVAQRCKELGITAMHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTP 133
Query 159 IPTDST 164
IPTDST
Sbjct 134 IPTDST 139
> At2g36160
Length=150
Score = 228 bits (581), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 107/127 (84%), Positives = 116/127 (91%), Gaps = 0/127 (0%)
Query 38 AAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYA 97
LGP RE E VFGV HI+ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYA
Sbjct 13 VVTLGPSVREGEQVFGVVHIFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYA 72
Query 98 AMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVT 157
AM+AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVT
Sbjct 73 AMLAAQDVAQRCKELGITAMHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVT 132
Query 158 PIPTDST 164
PIPTDST
Sbjct 133 PIPTDST 139
> At3g52580
Length=150
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 106/124 (85%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 41 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 100
LGP RE E VFGV H++ASFNDTFIHVTDLSGRETLVR+TGGMKVKADRDESSPYAAM+
Sbjct 16 LGPAVREGEQVFGVVHVFASFNDTFIHVTDLSGRETLVRITGGMKVKADRDESSPYAAML 75
Query 101 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 160
AA DVA RCKELGITA+HV++RATGG K+KTPGPGAQSALRALARSG++IGRIEDVTPIP
Sbjct 76 AAQDVAQRCKELGITAIHVKLRATGGNKTKTPGPGAQSALRALARSGMKIGRIEDVTPIP 135
Query 161 TDST 164
TDST
Sbjct 136 TDST 139
> CE00821
Length=152
Score = 226 bits (575), Expect = 2e-59, Method: Compositional matrix adjust.
Identities = 104/140 (74%), Positives = 124/140 (88%), Gaps = 0/140 (0%)
Query 25 APAAAAAAGGAAAAAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGM 84
APA A A +LGPQ +E EL+FGVAHI+ASFNDTF+H+TD+SGRET+VRVTGGM
Sbjct 2 APARKGKAKEEQAVVSLGPQAKEGELIFGVAHIFASFNDTFVHITDISGRETIVRVTGGM 61
Query 85 KVKADRDESSPYAAMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALA 144
KVKADRDESSPYAAM+AA DVA RCK+LGI A+H+++RATGGT++KTPGPGAQSALRALA
Sbjct 62 KVKADRDESSPYAAMLAAQDVADRCKQLGINALHIKLRATGGTRTKTPGPGAQSALRALA 121
Query 145 RSGLRIGRIEDVTPIPTDST 164
R+G++IGRIEDVTPIP+D T
Sbjct 122 RAGMKIGRIEDVTPIPSDCT 141
> 7290855
Length=151
Score = 222 bits (566), Expect = 3e-58, Method: Compositional matrix adjust.
Identities = 101/124 (81%), Positives = 116/124 (93%), Gaps = 0/124 (0%)
Query 41 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 100
LGPQ R+ E+VFGVAHIYASFNDTF+HVTDLSGRET+ RVTGGMKVKADRDE+SPYAAM+
Sbjct 17 LGPQVRDGEIVFGVAHIYASFNDTFVHVTDLSGRETIARVTGGMKVKADRDEASPYAAML 76
Query 101 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 160
AA DVA +CK LGITA+H+++RATGG K+KTPGPGAQSALRALARS ++IGRIEDVTPIP
Sbjct 77 AAQDVAEKCKTLGITALHIKLRATGGNKTKTPGPGAQSALRALARSSMKIGRIEDVTPIP 136
Query 161 TDST 164
+DST
Sbjct 137 SDST 140
> Hs5032051
Length=151
Score = 221 bits (563), Expect = 7e-58, Method: Compositional matrix adjust.
Identities = 101/125 (80%), Positives = 116/125 (92%), Gaps = 0/125 (0%)
Query 40 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 99
+LGPQ E E VFGV HI+ASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM
Sbjct 16 SLGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKETICRVTGGMKVKADRDESSPYAAM 75
Query 100 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPI 159
+AA DVA RCKELGITA+H+++RATGG ++KTPGPGAQSALRALARSG++IGRIEDVTPI
Sbjct 76 LAAQDVAQRCKELGITALHIKLRATGGNRTKTPGPGAQSALRALARSGMKIGRIEDVTPI 135
Query 160 PTDST 164
P+DST
Sbjct 136 PSDST 140
> YCR031c
Length=137
Score = 214 bits (546), Expect = 6e-56, Method: Compositional matrix adjust.
Identities = 99/121 (81%), Positives = 114/121 (94%), Gaps = 0/121 (0%)
Query 44 QTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAG 103
Q R++ VFGVA IYASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM+AA
Sbjct 6 QARDNSQVFGVARIYASFNDTFVHVTDLSGKETIARVTGGMKVKADRDESSPYAAMLAAQ 65
Query 104 DVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDS 163
DVAA+CKE+GITAVHV++RATGGT++KTPGPG Q+ALRALARSGLRIGRIEDVTP+P+DS
Sbjct 66 DVAAKCKEVGITAVHVKIRATGGTRTKTPGPGGQAALRALARSGLRIGRIEDVTPVPSDS 125
Query 164 T 164
T
Sbjct 126 T 126
> YJL191w
Length=138
Score = 214 bits (545), Expect = 7e-56, Method: Compositional matrix adjust.
Identities = 99/121 (81%), Positives = 114/121 (94%), Gaps = 0/121 (0%)
Query 44 QTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAG 103
Q R++ VFGVA IYASFNDTF+HVTDLSG+ET+ RVTGGMKVKADRDESSPYAAM+AA
Sbjct 7 QARDNSQVFGVARIYASFNDTFVHVTDLSGKETIARVTGGMKVKADRDESSPYAAMLAAQ 66
Query 104 DVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDS 163
DVAA+CKE+GITAVHV++RATGGT++KTPGPG Q+ALRALARSGLRIGRIEDVTP+P+DS
Sbjct 67 DVAAKCKEVGITAVHVKIRATGGTRTKTPGPGGQAALRALARSGLRIGRIEDVTPVPSDS 126
Query 164 T 164
T
Sbjct 127 T 127
> Hs20533883
Length=170
Score = 212 bits (540), Expect = 3e-55, Method: Compositional matrix adjust.
Identities = 97/127 (76%), Positives = 112/127 (88%), Gaps = 0/127 (0%)
Query 41 LGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMM 100
LGPQ E E VFGV HI+ASFNDTF+HVTDLSG++T+ RVTGGMKVKADRDESSPYAAM+
Sbjct 17 LGPQVAEGENVFGVCHIFASFNDTFVHVTDLSGKQTICRVTGGMKVKADRDESSPYAAML 76
Query 101 AAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIP 160
DVA RCKELGI A+H+++RATGG ++KT GPGAQSALRALA SG++IGRIEDVTPIP
Sbjct 77 TTQDVAQRCKELGIIALHIQLRATGGNRTKTLGPGAQSALRALACSGMKIGRIEDVTPIP 136
Query 161 TDSTRRK 167
+DST RK
Sbjct 137 SDSTLRK 143
> SPBC18H10.13
Length=139
Score = 184 bits (466), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 100/127 (78%), Positives = 118/127 (92%), Gaps = 0/127 (0%)
Query 38 AAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYA 97
A +GPQ R ELVFGVAHI+ASFNDTF+H+TDL+G+ET+VRVTGGMKVK DRDESSPYA
Sbjct 2 ATNVGPQIRSGELVFGVAHIFASFNDTFVHITDLTGKETIVRVTGGMKVKTDRDESSPYA 61
Query 98 AMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVT 157
AM+AA D AA+CKE+GITA+H+++RATGGT +KTPGPGAQ+ALRALAR+G+RIGRIEDVT
Sbjct 62 AMLAAQDAAAKCKEVGITALHIKIRATGGTATKTPGPGAQAALRALARAGMRIGRIEDVT 121
Query 158 PIPTDST 164
PIPTDST
Sbjct 122 PIPTDST 128
> SPAC3H5.05c
Length=139
Score = 184 bits (466), Expect = 1e-46, Method: Compositional matrix adjust.
Identities = 100/127 (78%), Positives = 118/127 (92%), Gaps = 0/127 (0%)
Query 38 AAALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYA 97
A +GPQ R ELVFGVAHI+ASFNDTF+H+TDL+G+ET+VRVTGGMKVK DRDESSPYA
Sbjct 2 ATNVGPQIRSGELVFGVAHIFASFNDTFVHITDLTGKETIVRVTGGMKVKTDRDESSPYA 61
Query 98 AMMAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVT 157
AM+AA D AA+CKE+GITA+H+++RATGGT +KTPGPGAQ+ALRALAR+G+RIGRIEDVT
Sbjct 62 AMLAAQDAAAKCKEVGITALHIKIRATGGTATKTPGPGAQAALRALARAGMRIGRIEDVT 121
Query 158 PIPTDST 164
PIPTDST
Sbjct 122 PIPTDST 128
> ECU03g0650
Length=134
Score = 138 bits (348), Expect = 5e-33, Method: Compositional matrix adjust.
Identities = 63/109 (57%), Positives = 83/109 (76%), Gaps = 0/109 (0%)
Query 54 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 113
VAHI A+ NDTF+H+TD++G ET+ ++TGGM+VKA RDE SPYAAM+AA DVA + G
Sbjct 13 VAHIKATKNDTFVHITDMTGSETIAKITGGMRVKAQRDEGSPYAAMLAAQDVATKILGRG 72
Query 114 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTD 162
+ +H ++R GG K GPGAQ+A+R L R+GLR+GRIEDVTP+ D
Sbjct 73 VKVLHFKLRGAGGVKPMALGPGAQTAIRTLIRAGLRVGRIEDVTPVARD 121
> Hs22066787
Length=224
Score = 72.4 bits (176), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 44/111 (39%), Positives = 54/111 (48%), Gaps = 40/111 (36%)
Query 40 ALGPQTREDELVFGVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAM 99
+LGP E VFGV I+ASF DT
Sbjct 16 SLGPWVGVGENVFGVCQIFASFTDTL---------------------------------- 41
Query 100 MAAGDVAARCKELGITAVHVRVRATGGTKSKTPGPGAQSALRA--LARSGL 148
DVA RCKELGITA+H+++ ATGG ++K+P PGAQ +LRA L RS L
Sbjct 42 ----DVAQRCKELGITALHIKLWATGGNRNKSPRPGAQMSLRAPCLLRSWL 88
> AtCh055
Length=138
Score = 66.2 bits (160), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 43/119 (36%), Positives = 61/119 (51%), Gaps = 9/119 (7%)
Query 53 GVAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKEL 112
GV H+ ASFN+T + VTD+ GR G + R +P+AA AAG+ +
Sbjct 28 GVIHVQASFNNTIVTVTDVRGRVISWSSAGTCGFRGTR-RGTPFAAQTAAGNAIRAVVDQ 86
Query 113 GITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 171
G+ VR+ K PG G +ALRA+ RSG+ + + DVTP+P + R RR
Sbjct 87 GMQRAEVRI--------KGPGLGRDAALRAIRRSGILLSFVRDVTPMPHNGCRPPKKRR 137
> 7300221
Length=200
Score = 45.1 bits (105), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 38/117 (32%), Positives = 53/117 (45%), Gaps = 9/117 (7%)
Query 54 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 113
+ +I S N+T I VTD G L+R G K R ++ AA A ++ + ELG
Sbjct 91 ICNIRVSPNNTIISVTDHKGVLRLIRSCGIEGFKNTRKGTN-IAAQATAVTISGKAIELG 149
Query 114 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGR 170
V V+VR GPG SA++ L GL I I D T + + R + R
Sbjct 150 WKTVRVKVRGL--------GPGRMSAIKGLQMGGLNIVSITDNTHVSFNPPRPRKQR 198
> Hs16554609
Length=194
Score = 42.7 bits (99), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 41/118 (34%), Positives = 57/118 (48%), Gaps = 9/118 (7%)
Query 54 VAHIYASFNDTFIHVTDLSGRETLVRVTGGMKVKADRDESSPYAAMMAAGDVAARCKELG 113
+AHI AS N+T I V S E L + G + + + + AA A AAR K+ G
Sbjct 85 IAHIKASHNNTQIQVVSASN-EPLAFASCGTEGFRNAKKGTGIAAQTAGIAAAARAKQKG 143
Query 114 ITAVHVRVRATGGTKSKTPGPGAQSALRALARSGLRIGRIEDVTPIPTDSTRRKSGRR 171
+ +H+RV G GPG SA+ L GL + I D TPIP + R + R+
Sbjct 144 V--IHIRVVVKGL------GPGRLSAMHGLIMGGLEVISITDNTPIPHNGCRPRKARK 193
> At1g31817
Length=235
Score = 30.8 bits (68), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 35/69 (50%), Gaps = 2/69 (2%)
Query 54 VAHIYASFNDTFIHVTDLSGRETLVRVTGGM-KVKADRDESSPYAAMMAAGDVAARCKEL 112
+ HI N+TF+ VTD G +G + +K R ++ Y A A ++ R K +
Sbjct 116 IIHIKMLRNNTFVTVTDSKGNVKCKATSGSLPDLKGGRKMTN-YTADATAENIGRRAKAM 174
Query 113 GITAVHVRV 121
G+ +V V+V
Sbjct 175 GLKSVVVKV 183
Lambda K H
0.314 0.128 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2671071884
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40