bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3513_orf6
Length=209
Score E
Sequences producing significant alignments: (Bits) Value
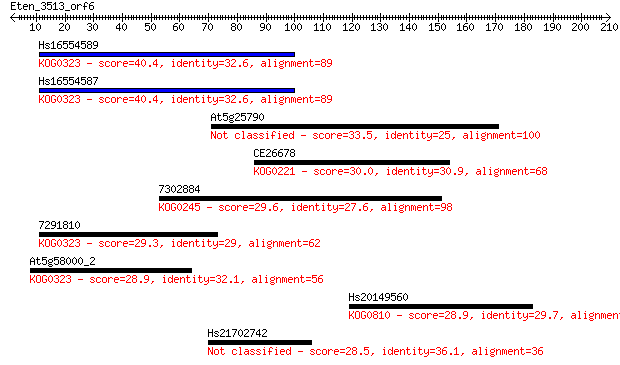
Hs16554589 40.4 0.002
Hs16554587 40.4 0.003
At5g25790 33.5 0.34
CE26678 30.0 3.8
7302884 29.6 5.0
7291810 29.3 5.1
At5g58000_2 28.9 7.1
Hs20149560 28.9 7.9
Hs21702742 28.5 9.6
> Hs16554589
Length=867
Score = 40.4 bits (93), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 42/91 (46%), Gaps = 4/91 (4%)
Query 11 VTHVMAGKSNTNNMLALKERSFQHLKKVHTLWLFACESMWAKAPESCF--DADALCALYD 68
TH++A ++ T +L +E HL V+ WL++C W K E F D A +
Sbjct 681 ATHLIAARAGTEKVLQAQE--CGHLHVVNPDWLWSCLERWDKVEEQLFPLRDDHTKAQRE 738
Query 69 NQPPCAPFKDHWMHLAEFVPPPAVAPAQPLP 99
N P P ++ A F P P + AQP P
Sbjct 739 NSPAAFPDREGVPPTALFHPMPVLPKAQPGP 769
> Hs16554587
Length=961
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 42/91 (46%), Gaps = 4/91 (4%)
Query 11 VTHVMAGKSNTNNMLALKERSFQHLKKVHTLWLFACESMWAKAPESCF--DADALCALYD 68
TH++A ++ T +L +E HL V+ WL++C W K E F D A +
Sbjct 681 ATHLIAARAGTEKVLQAQE--CGHLHVVNPDWLWSCLERWDKVEEQLFPLRDDHTKAQRE 738
Query 69 NQPPCAPFKDHWMHLAEFVPPPAVAPAQPLP 99
N P P ++ A F P P + AQP P
Sbjct 739 NSPAAFPDREGVPPTALFHPMPVLPKAQPGP 769
> At5g25790
Length=408
Score = 33.5 bits (75), Expect = 0.34, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 42/106 (39%), Gaps = 12/106 (11%)
Query 71 PPCAPFKDHWMHLAEFVP---PPAVAPAQPLPLQDRLPVREFLGTGPYSDGASLISPFEE 127
PPC+P K E P PP ++R+ E +G Y +G + ++
Sbjct 33 PPCSPVKSSQQSETEVTPPQKPPLHQFGHNQVQKNRVRYCECFASGSYCNGCNCVNCHN- 91
Query 128 TIFLWRPEKQPIRQLYSKASTQHSPPTLA---AGSPHPQQQLQQQL 170
+ E + RQ+ + +P AGSPH + LQ+ +
Sbjct 92 -----KLENESSRQVAISGILERNPDAFKPKIAGSPHGMKDLQENV 132
> CE26678
Length=1369
Score = 30.0 bits (66), Expect = 3.8, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 28/68 (41%), Gaps = 0/68 (0%)
Query 86 FVPPPAVAPAQPLPLQDRLPVREFLGTGPYSDGASLISPFEETIFLWRPEKQPIRQLYSK 145
F P V +Q L +L + FL S +IS E P K PI S+
Sbjct 1043 FKKTPNVKESQVLETPKQLSISSFLEPKFPSSEKDVISRVSERYLQSDPFKTPISDRRSQ 1102
Query 146 ASTQHSPP 153
S++HS P
Sbjct 1103 QSSRHSTP 1110
> 7302884
Length=1773
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 27/117 (23%), Positives = 44/117 (37%), Gaps = 26/117 (22%)
Query 53 APESCFDADALCALYDNQPPCAPFK----DHWMHLAEFVPPPAVAPAQPLPLQDRLPVRE 108
+ E C D C ++ C+P + + W P PA PA LPL+ +P E
Sbjct 1581 SDEGCADMTVSCISSNSMELCSPDRADAPNGWE-----APAPATQPA--LPLRLYVPELE 1633
Query 109 FLGTGPYSDGASLISPFEET---------------IFLWRPEKQPIRQLYSKASTQH 150
+ P L++ E +F++R EK P+ + +T H
Sbjct 1634 EIRVSPVVARKGLLNVLEHGGSGWKKRWVIVRRPYVFIYRSEKDPVERAVLNLATAH 1690
> 7291810
Length=880
Score = 29.3 bits (64), Expect = 5.1, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 2/62 (3%)
Query 11 VTHVMAGKSNTNNMLALKERSFQHLKKVHTLWLFACESMWAKAPESCFDADALCALYDNQ 70
+TH++A + T + A K+ +K V+ WL+ C W E F D Q
Sbjct 632 ITHLVAVNAGTYKVNAAKKEP--AIKVVNANWLWTCAERWEHVEEKLFPLDRKVRNKGRQ 689
Query 71 PP 72
PP
Sbjct 690 PP 691
> At5g58000_2
Length=374
Score = 28.9 bits (63), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 18/57 (31%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query 8 EPGVTHVMAGKSNTNNML-ALKERSFQHLKKVHTLWLFACESMWAKAPESCFDADAL 63
+ VTHV+A T A++E+ + VH W+ A +W K PE F + L
Sbjct 314 DASVTHVVAMDVGTEKARWAVREKKYV----VHRGWIDAANYLWMKQPEENFGLEQL 366
> Hs20149560
Length=297
Score = 28.9 bits (63), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 119 ASLISPFEETIFLWRPEKQPIRQLYSKASTQHSPPTLAAGSPHPQQQLQQQLLPLRPEGQ 178
A L SP EE R +Q I +L +K +P P++ ++Q+L LR E +
Sbjct 32 ARLGSPDEEFFHKVRTIRQTIVKLGNKVQELEKQQVTILATPLPEESMKQELQNLRDEIK 91
Query 179 NVGQ 182
+G+
Sbjct 92 QLGR 95
> Hs21702742
Length=1709
Score = 28.5 bits (62), Expect = 9.6, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 17/36 (47%), Gaps = 0/36 (0%)
Query 70 QPPCAPFKDHWMHLAEFVPPPAVAPAQPLPLQDRLP 105
QPP P +L VPPP ++P P+ L P
Sbjct 1096 QPPAQPLSSSQPNLRAQVPPPLLSPQVPVSLLKYAP 1131
Lambda K H
0.319 0.134 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3754464630
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40