bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3564_orf1
Length=233
Score E
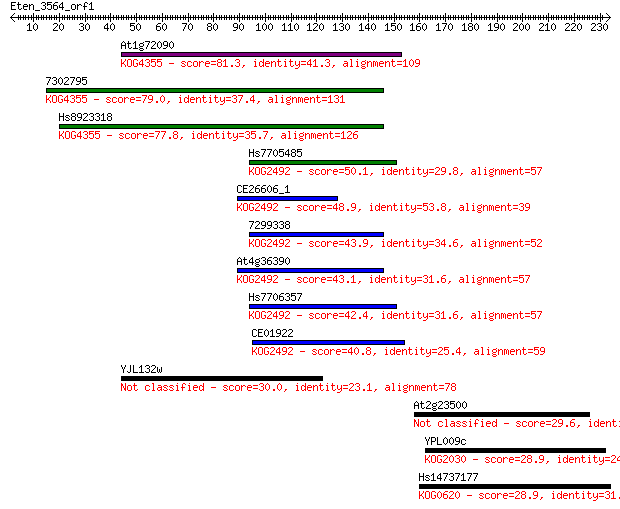
Sequences producing significant alignments: (Bits) Value
At1g72090 81.3 2e-15
7302795 79.0 8e-15
Hs8923318 77.8 2e-14
Hs7705485 50.1 4e-06
CE26606_1 48.9 9e-06
7299338 43.9 3e-04
At4g36390 43.1 5e-04
Hs7706357 42.4 9e-04
CE01922 40.8 0.002
YJL132w 30.0 4.6
At2g23500 29.6 5.5
YPL009c 28.9 8.2
Hs14737177 28.9 8.3
> At1g72090
Length=601
Score = 81.3 bits (199), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 45/109 (41%), Positives = 65/109 (59%), Gaps = 13/109 (11%)
Query 44 IQPSTTRRRTTKQMDSEGNQTSDRDNSTTATTPPSSATHAVEGAFLPGRAKIFFKTFGCS 103
I P T + +K++ S+ +Q + + + A PPS +PG I+ KTFGCS
Sbjct 28 INPKTNK---SKRISSKPDQITASNRDSLA--PPSMK--------IPGTQTIYIKTFGCS 74
Query 104 HNISDSEYMQGLLHESGYRFVDSLEEADICVVNSCTVKSPSEFALYSVI 152
HN SDSEYM G L GY + EEAD+ ++N+CTVKSPS+ A+ ++I
Sbjct 75 HNQSDSEYMAGQLSAFGYALTEVPEEADLWLINTCTVKSPSQSAMSTLI 123
> 7302795
Length=552
Score = 79.0 bits (193), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 49/133 (36%), Positives = 71/133 (53%), Gaps = 19/133 (14%)
Query 15 DISGTDLEDLRS--SDDDMKRAARVTGLYTYIQPSTTRRRTTKQMDSEGNQTSDRDNSTT 72
D+ G D++D+ S DD+K R Y + T R + Q+ E + ++ T
Sbjct 7 DLPGNDVDDIEDLISADDVKPRER----YENKKTVTVRAKKRSQIRLESQEEEEKPKPT- 61
Query 73 ATTPPSSATHAVEGAFLPGRAKIFFKTFGCSHNISDSEYMQGLLHESGYRFVDSLEEADI 132
+ + +PG K+F KT+GC+HN SDSEYM G L GYR + EEAD+
Sbjct 62 -----------IHESVIPGTQKVFVKTWGCAHNNSDSEYMAGQLAAYGYR-LSGKEEADL 109
Query 133 CVVNSCTVKSPSE 145
++NSCTVK+PSE
Sbjct 110 WLLNSCTVKNPSE 122
> Hs8923318
Length=579
Score = 77.8 bits (190), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 45/126 (35%), Positives = 67/126 (53%), Gaps = 21/126 (16%)
Query 20 DLEDLRSSDDDMKRAARVTGLYTYIQPSTTRRRTTKQMDSEGNQTSDRDNSTTATTPPSS 79
D+ED+ S +D + + + P RR T K + E N +PPS
Sbjct 12 DIEDIVSQEDSKPQDRHF--VRKDVVPKVRRRNTQKYLQEEEN------------SPPSD 57
Query 80 ATHAVEGAFLPGRAKIFFKTFGCSHNISDSEYMQGLLHESGYRFVDSLEEADICVVNSCT 139
+T +PG KI+ +T+GCSHN SD EYM G L GY+ ++ +AD+ ++NSCT
Sbjct 58 ST-------IPGIQKIWIRTWGCSHNNSDGEYMAGQLAAYGYKITENASDADLWLLNSCT 110
Query 140 VKSPSE 145
VK+P+E
Sbjct 111 VKNPAE 116
> Hs7705485
Length=586
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 39/57 (68%), Gaps = 0/57 (0%)
Query 94 KIFFKTFGCSHNISDSEYMQGLLHESGYRFVDSLEEADICVVNSCTVKSPSEFALYS 150
K++ +T+GC N++D+E +L +SGY +L+EAD+ ++ +C+++ +E +++
Sbjct 101 KVYLETYGCQMNVNDTEIAWSILQKSGYLRTSNLQEADVILLVTCSIREKAEQTIWN 157
> CE26606_1
Length=134
Score = 48.9 bits (115), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 21/40 (52%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query 89 LPGRA-KIFFKTFGCSHNISDSEYMQGLLHESGYRFVDSL 127
+PG K++ +T+GCSHN SDSEYM GLL ++GY V
Sbjct 43 VPGVGQKVWVRTWGCSHNTSDSEYMSGLLQQAGYDVVKGF 82
> 7299338
Length=583
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 32/52 (61%), Gaps = 0/52 (0%)
Query 94 KIFFKTFGCSHNISDSEYMQGLLHESGYRFVDSLEEADICVVNSCTVKSPSE 145
K+ F+ +GC N +D+E + +L E+GY EEAD+ ++ +C V+ +E
Sbjct 93 KVHFEVYGCQMNTNDTEVVFSILKENGYLRCQEPEEADVIMLVTCAVRDGAE 144
> At4g36390
Length=587
Score = 43.1 bits (100), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 35/58 (60%), Gaps = 1/58 (1%)
Query 89 LPGRAKIFFKTFGCSHNISDSEYMQGLLHESGYR-FVDSLEEADICVVNSCTVKSPSE 145
+ + +I+ +T+GC NI+D E + ++ SGY+ V E A++ VN+C ++ +E
Sbjct 72 IASKGRIYHETYGCQMNINDMEIVLAIMKNSGYKEVVTDPESAEVIFVNTCAIRENAE 129
> Hs7706357
Length=554
Score = 42.4 bits (98), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 38/58 (65%), Gaps = 3/58 (5%)
Query 94 KIFFKTFGCSHNISDSEYMQGLLHESGY-RFVDSLEEADICVVNSCTVKSPSEFALYS 150
K++ +T+GC N++D+E +L +SGY R V S AD+ ++ +C+++ +E +++
Sbjct 62 KVYLETYGCQMNVNDTEIAWSILQKSGYLRPVTS--RADVILLVTCSIREKAEQTIWN 117
> CE01922
Length=547
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 15/59 (25%), Positives = 37/59 (62%), Gaps = 0/59 (0%)
Query 95 IFFKTFGCSHNISDSEYMQGLLHESGYRFVDSLEEADICVVNSCTVKSPSEFALYSVIR 153
+ + T+GC N+SD E ++ ++ + G+ D E ADI ++ +C+++ +E +++ ++
Sbjct 81 VCYVTYGCQMNVSDMEIVRSIMTKYGFVESDKKENADIVLLMTCSIRDGAEKKVWNQLK 139
> YJL132w
Length=750
Score = 30.0 bits (66), Expect = 4.6, Method: Composition-based stats.
Identities = 18/78 (23%), Positives = 31/78 (39%), Gaps = 0/78 (0%)
Query 44 IQPSTTRRRTTKQMDSEGNQTSDRDNSTTATTPPSSATHAVEGAFLPGRAKIFFKTFGCS 103
I+ TT Q GN D+ ++P A F+PG + I + T
Sbjct 431 IKDETTEEAHQLQFAVTGNFYDDKIPDLVVSSPSYGANETGIATFIPGSSIISYLTNSDK 490
Query 104 HNISDSEYMQGLLHESGY 121
+ + D +G+++ GY
Sbjct 491 YQVVDISTFKGVINLDGY 508
> At2g23500
Length=784
Score = 29.6 bits (65), Expect = 5.5, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 34/68 (50%), Gaps = 4/68 (5%)
Query 158 EIPNNVETTGTSSTRQQGRNAYSSQSIMDNAHRTNDSGTHSLVEQQQKQQHEEQRQREEA 217
++P + G SS G+N Y+ + ++ R +D L+E++Q QH+ + E+
Sbjct 100 QLPEQLSRVGNSSV---GKN-YTELNSEEDEMRVDDGALIVLLEEEQGTQHQLEAIVEDH 155
Query 218 DPQRGQTR 225
Q +TR
Sbjct 156 GTQEDETR 163
> YPL009c
Length=1038
Score = 28.9 bits (63), Expect = 8.2, Method: Composition-based stats.
Identities = 17/70 (24%), Positives = 33/70 (47%), Gaps = 0/70 (0%)
Query 162 NVETTGTSSTRQQGRNAYSSQSIMDNAHRTNDSGTHSLVEQQQKQQHEEQRQREEADPQR 221
N G ++ + Y+ Q + R GT +E+QQ+++ EE +RE + ++
Sbjct 834 NSNVRGKRGKLKKIQKKYADQDETERLLRLEALGTLKGIEKQQQRKKEEIMKREVREDRK 893
Query 222 GQTRTTRRLK 231
+ RRL+
Sbjct 894 NKREKQRRLQ 903
> Hs14737177
Length=530
Score = 28.9 bits (63), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 0/74 (0%)
Query 160 PNNVETTGTSSTRQQGRNAYSSQSIMDNAHRTNDSGTHSLVEQQQKQQHEEQRQREEADP 219
P +V T+S+R + S + A R +S +L E Q+KQ EQ++RE+
Sbjct 304 PRHVFRRKTASSRSILPDLLSPYQMAIRAKRLEESRAAALRELQEKQALMEQQRREKRAL 363
Query 220 QRGQTRTTRRLKRR 233
Q + R R KR+
Sbjct 364 QEWRERAQRMRKRK 377
Lambda K H
0.309 0.124 0.341
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4562417408
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40