bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
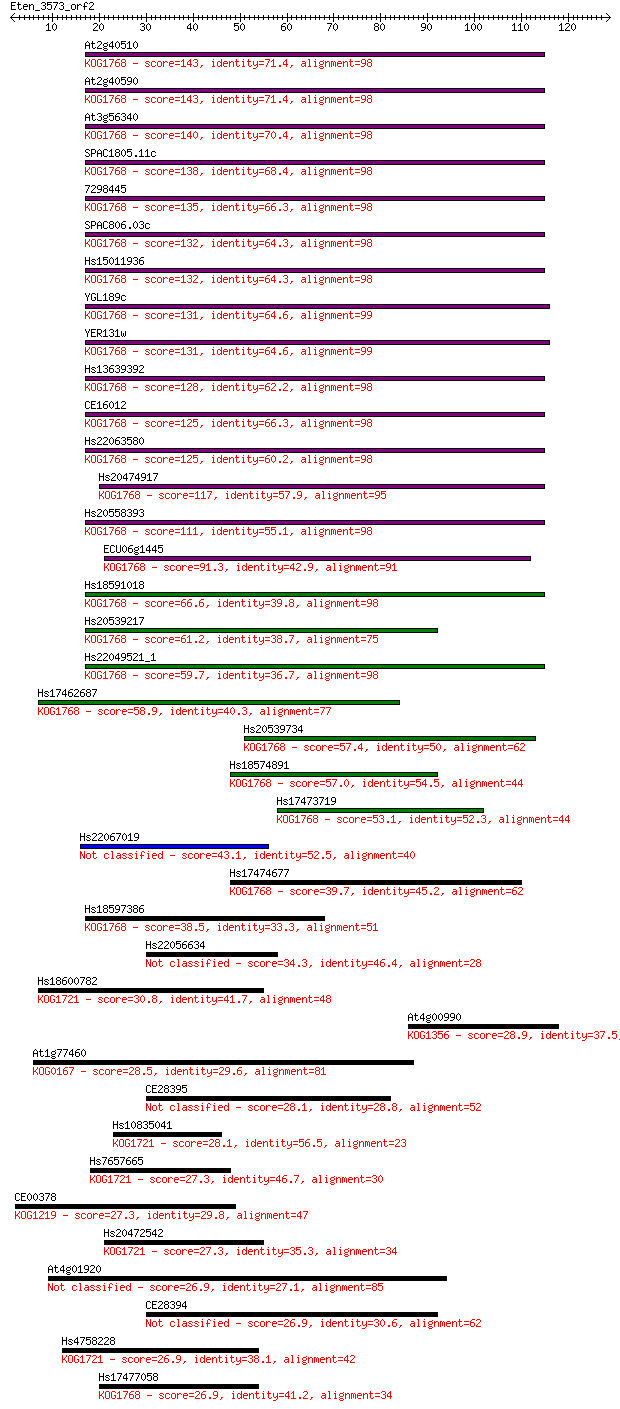
Query= Eten_3573_orf2
Length=128
Score E
Sequences producing significant alignments: (Bits) Value
At2g40510 143 9e-35
At2g40590 143 1e-34
At3g56340 140 4e-34
SPAC1805.11c 138 2e-33
7298445 135 2e-32
SPAC806.03c 132 1e-31
Hs15011936 132 2e-31
YGL189c 131 3e-31
YER131w 131 4e-31
Hs13639392 128 3e-30
CE16012 125 1e-29
Hs22063580 125 3e-29
Hs20474917 117 7e-27
Hs20558393 111 3e-25
ECU06g1445 91.3 4e-19
Hs18591018 66.6 1e-11
Hs20539217 61.2 4e-10
Hs22049521_1 59.7 1e-09
Hs17462687 58.9 2e-09
Hs20539734 57.4 7e-09
Hs18574891 57.0 9e-09
Hs17473719 53.1 1e-07
Hs22067019 43.1 1e-04
Hs17474677 39.7 0.001
Hs18597386 38.5 0.004
Hs22056634 34.3 0.053
Hs18600782 30.8 0.69
At4g00990 28.9 2.5
At1g77460 28.5 3.1
CE28395 28.1 4.0
Hs10835041 28.1 4.1
Hs7657665 27.3 6.6
CE00378 27.3 7.8
Hs20472542 27.3 8.2
At4g01920 26.9 9.0
CE28394 26.9 9.4
Hs4758228 26.9 9.4
Hs17477058 26.9 9.9
> At2g40510
Length=133
Score = 143 bits (360), Expect = 9e-35, Method: Compositional matrix adjust.
Identities = 70/98 (71%), Positives = 79/98 (80%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KH RGHVNP+RCSNCG+ CPKDKAIKRF VRNIV+ ++ RD++EAS Y
Sbjct 1 MTFKRRNGGRNKHNRGHVNPIRCSNCGKCCPKDKAIKRFIVRNIVEQAAIRDVQEASVYE 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY K YC+SCAIHS VVRVRSR RRVRT P
Sbjct 61 GYTLPKLYAKTQYCVSCAIHSHVVRVRSRTNRRVRTPP 98
> At2g40590
Length=131
Score = 143 bits (360), Expect = 1e-34, Method: Compositional matrix adjust.
Identities = 70/98 (71%), Positives = 79/98 (80%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KH RGHVNP+RCSNCG+ CPKDKAIKRF VRNIV+ ++ RD++EAS Y
Sbjct 1 MTFKRRNGGRNKHNRGHVNPIRCSNCGKCCPKDKAIKRFIVRNIVEQAAIRDVQEASVYE 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY K YC+SCAIHS VVRVRSR RRVRT P
Sbjct 61 GYTLPKLYAKTQYCVSCAIHSHVVRVRSRTNRRVRTPP 98
> At3g56340
Length=130
Score = 140 bits (354), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 69/98 (70%), Positives = 78/98 (79%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KH RGHV P+RCSNCG+ CPKDKAIKRF VRNIV+ ++ RD++EAS Y
Sbjct 1 MTFKRRNGGRNKHNRGHVKPIRCSNCGKCCPKDKAIKRFIVRNIVEQAAIRDVQEASVYE 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY K YC+SCAIHS VVRVRSR RRVRT P
Sbjct 61 GYTLPKLYAKTQYCVSCAIHSHVVRVRSRTNRRVRTPP 98
> SPAC1805.11c
Length=119
Score = 138 bits (348), Expect = 2e-33, Method: Compositional matrix adjust.
Identities = 67/98 (68%), Positives = 79/98 (80%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KHGRGHV VRC NC R+ PKDKAIKR+++RN+V+ ++ RDL EAS Y
Sbjct 1 MTQKRRNNGRNKHGRGHVKFVRCINCSRAVPKDKAIKRWTIRNMVETAAIRDLSEASVYS 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y +PKLYIK YC+SCAIHSRVVRVRSRE RR+RT P
Sbjct 61 EYTIPKLYIKLQYCVSCAIHSRVVRVRSREGRRIRTPP 98
> 7298445
Length=114
Score = 135 bits (339), Expect = 2e-32, Method: Compositional matrix adjust.
Identities = 65/98 (66%), Positives = 78/98 (79%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KH RGHV PVRC+NC R PKDKAIK+F +RNIV+A++ RD+ EAS +
Sbjct 1 MTKKRRNGGRNKHNRGHVKPVRCTNCARCVPKDKAIKKFVIRNIVEAAAVRDITEASIWD 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
+Y LPKLY K YC+SCAIHS+VVR RSRE RR+RT P
Sbjct 61 SYVLPKLYAKLHYCVSCAIHSKVVRNRSREARRIRTPP 98
> SPAC806.03c
Length=120
Score = 132 bits (333), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 63/98 (64%), Positives = 78/98 (79%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+KHGRGH VRC NC R+ PKDKAIKR+++RN+V+ ++ RDL EAS Y
Sbjct 1 MTQKRRNCGRNKHGRGHTKFVRCINCSRAVPKDKAIKRWNIRNMVETAAIRDLSEASVYS 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y +PK+Y+K YC+SCAIH+RVVRVRSRE RR+RT P
Sbjct 61 EYAIPKIYVKLQYCVSCAIHARVVRVRSREGRRIRTPP 98
> Hs15011936
Length=115
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 63/98 (64%), Positives = 77/98 (78%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+K GRGHV P+RC+NC R PKDKAIK+F +RNIV+A++ RD+ EAS +
Sbjct 1 MTKKRRNNGRAKKGRGHVQPIRCTNCARCVPKDKAIKKFVIRNIVEAAAVRDISEASVFD 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY+K YC+SCAIHS+VVR RSRE R+ RT P
Sbjct 61 AYVLPKLYVKLHYCVSCAIHSKVVRNRSREARKDRTPP 98
> YGL189c
Length=119
Score = 131 bits (329), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 64/99 (64%), Positives = 76/99 (76%), Gaps = 0/99 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
MP KR + GR+K GRGHV PVRC NC +S PKDKAIKR ++RNIV+A++ RDL EAS Y
Sbjct 1 MPKKRASNGRNKKGRGHVKPVRCVNCSKSIPKDKAIKRMAIRNIVEAAAVRDLSEASVYP 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNPQ 115
Y LPK Y K YC+SCAIH+R+VRVRSRE R+ R PQ
Sbjct 61 EYALPKTYNKLHYCVSCAIHARIVRVRSREDRKNRAPPQ 99
> YER131w
Length=119
Score = 131 bits (329), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 64/99 (64%), Positives = 76/99 (76%), Gaps = 0/99 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
MP KR + GR+K GRGHV PVRC NC +S PKDKAIKR ++RNIV+A++ RDL EAS Y
Sbjct 1 MPKKRASNGRNKKGRGHVKPVRCVNCSKSIPKDKAIKRMAIRNIVEAAAVRDLSEASVYP 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNPQ 115
Y LPK Y K YC+SCAIH+R+VRVRSRE R+ R PQ
Sbjct 61 EYALPKTYNKLHYCVSCAIHARIVRVRSREDRKNRAPPQ 99
> Hs13639392
Length=115
Score = 128 bits (321), Expect = 3e-30, Method: Compositional matrix adjust.
Identities = 61/98 (62%), Positives = 75/98 (76%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+K GRGHV P+RC+NC R PKDKAIK+ +RNIV+A++ RD+ EAS +
Sbjct 1 MTKKRRNNGRAKKGRGHVQPIRCTNCARCVPKDKAIKKLVIRNIVEATAVRDISEASVFD 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY+K YC+SCAIHS+VVR RS E R+ RT P
Sbjct 61 AYVLPKLYVKLHYCVSCAIHSKVVRNRSHEARKDRTPP 98
> CE16012
Length=117
Score = 125 bits (315), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 65/98 (66%), Positives = 76/98 (77%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+K RGHV +RC+NCGR CPKDKAIK+F VRNIV+A++ RD+ +ASAY
Sbjct 1 MTFKRRNHGRNKKNRGHVAFIRCTNCGRCCPKDKAIKKFVVRNIVEAAAVRDIGDASAYT 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY K YCI+CAIHS+VVR RSRE RR R P
Sbjct 61 QYALPKLYHKLHYCIACAIHSKVVRNRSREARRDRNPP 98
> Hs22063580
Length=115
Score = 125 bits (313), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 59/98 (60%), Positives = 74/98 (75%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN GR+K GRGH+ P+R +NC R PKDK IK+F +RNIV+A++ RD+ E S +
Sbjct 1 MTKKRRNNGRAKKGRGHLQPIRWTNCARCVPKDKGIKKFVIRNIVEAATVRDISEVSVFD 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY+K YC+SCAIHS+VVR RSRE R+ RT P
Sbjct 61 AYVLPKLYVKLHYCVSCAIHSKVVRNRSREARKDRTPP 98
> Hs20474917
Length=153
Score = 117 bits (292), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 55/95 (57%), Positives = 72/95 (75%), Gaps = 0/95 (0%)
Query 20 KRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYHTYN 79
KRRN GR+K G GHV P+ C+NC + PKDKAIK+F +RNIV+A++ RD+ EA + Y
Sbjct 42 KRRNNGRAKKGHGHVQPICCTNCAQCVPKDKAIKKFVIRNIVEAAAVRDISEAGIFDAYV 101
Query 80 LPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
LPKLY+K YC+SCAIH++VVR +S E R+ RT P
Sbjct 102 LPKLYVKLHYCVSCAIHNKVVRNQSCEARKDRTPP 136
> Hs20558393
Length=115
Score = 111 bits (277), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KR N GR+K GRGH+ P+RC NC R PKDKAIK+F + NIV+A++ RD+ EAS +
Sbjct 1 MTKKRGNNGRAKKGRGHMQPIRCRNCARCIPKDKAIKKFIIHNIVEAAAVRDISEASVFD 60
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY+K Y +SCA+HS+VVR +S R+ +T P
Sbjct 61 AYVLPKLYVKLHYFLSCAVHSKVVRNQSCVARKDQTPP 98
> ECU06g1445
Length=105
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 39/91 (42%), Positives = 63/91 (69%), Gaps = 0/91 (0%)
Query 21 RRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYHTYNL 80
RRN GR K RG ++C CG PKDKAIKRF ++++++ +S DL++A+ Y + +
Sbjct 2 RRNHGRGKKNRGSAGSIQCDKCGSVTPKDKAIKRFRIQSLIEQASFDDLKQATIYDVFEV 61
Query 81 PKLYIKHCYCISCAIHSRVVRVRSRERRRVR 111
P++ K +C+SCA H+++VRVRS + R++R
Sbjct 62 PRMGYKSQFCVSCACHAKIVRVRSSQARKIR 92
> Hs18591018
Length=191
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/98 (39%), Positives = 49/98 (50%), Gaps = 27/98 (27%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KR+N +K GRGH+ P+ C NC R PK KA+K+
Sbjct 1 MTKKRKNNNHAKKGRGHMQPICCMNCARCMPKYKAVKKL--------------------- 39
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
LY+K YC+SCAIHS+VVR RS E R+ T P
Sbjct 40 ------LYVKLHYCVSCAIHSKVVRNRSHEARKDPTPP 71
> Hs20539217
Length=141
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/75 (38%), Positives = 42/75 (56%), Gaps = 13/75 (17%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN ++ GR HV + C N + + NI +A++ RD+ EAS ++
Sbjct 1 MTKKRRNNDYAQKGREHVQSIHCMN-------------YIILNIAEATAVRDISEASVFN 47
Query 77 TYNLPKLYIKHCYCI 91
Y LPKLY+K CYC+
Sbjct 48 AYVLPKLYLKLCYCM 62
> Hs22049521_1
Length=89
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 36/98 (36%), Positives = 45/98 (45%), Gaps = 30/98 (30%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYH 76
M KRRN G +K G HV P+RC+N AS +
Sbjct 1 MTKKRRNNGHAKKGHSHVQPIRCTN------------------------------ASVFD 30
Query 77 TYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRVRTNP 114
Y LPKLY+K C+ CA+HS+VVR S E + RT P
Sbjct 31 AYVLPKLYVKLLSCVGCAMHSKVVRNGSHEAHKDRTPP 68
> Hs17462687
Length=133
Score = 58.9 bits (141), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/77 (40%), Positives = 46/77 (59%), Gaps = 1/77 (1%)
Query 7 SGLRRQEAERMPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQ 66
+ L R + KRRN +K G GHV P C+NC + PKDK IK FS++ IV+A++
Sbjct 28 ASLSRVLTSKKTKKRRNNDGAKKGCGHVQPFHCTNCIQCVPKDKVIK-FSIQGIVEATTV 86
Query 67 RDLREASAYHTYNLPKL 83
RD+ ++ + + L KL
Sbjct 87 RDISKSRIFAAHVLLKL 103
> Hs20539734
Length=116
Score = 57.4 bits (137), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 31/62 (50%), Positives = 44/62 (70%), Gaps = 0/62 (0%)
Query 51 AIKRFSVRNIVDASSQRDLREASAYHTYNLPKLYIKHCYCISCAIHSRVVRVRSRERRRV 110
AIK+F +RN V+A++ RD+ + S +++ LPKLY K +SCAIHS+VVR RS E +
Sbjct 4 AIKKFVIRNTVEATAVRDISKVSIFNSEVLPKLYGKLRCAVSCAIHSKVVRNRSCEACKD 63
Query 111 RT 112
RT
Sbjct 64 RT 65
> Hs18574891
Length=107
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 24/44 (54%), Positives = 34/44 (77%), Gaps = 0/44 (0%)
Query 48 KDKAIKRFSVRNIVDASSQRDLREASAYHTYNLPKLYIKHCYCI 91
KDKAIK+F ++NIV+A++ RD+ EAS + Y PKLY+K YC+
Sbjct 52 KDKAIKKFVIQNIVEATAVRDISEASVFDAYVRPKLYVKLHYCV 95
> Hs17473719
Length=105
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 23/44 (52%), Positives = 33/44 (75%), Gaps = 0/44 (0%)
Query 58 RNIVDASSQRDLREASAYHTYNLPKLYIKHCYCISCAIHSRVVR 101
+N V+A++ RD+ E S + Y LPKLY+K +C+ CAIHS+VVR
Sbjct 61 KNQVEAATVRDISEISIFDAYVLPKLYVKLHHCVGCAIHSKVVR 104
> Hs22067019
Length=162
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 21/46 (45%), Positives = 27/46 (58%), Gaps = 6/46 (13%)
Query 16 RMPTKRRNAGRSK------HGRGHVNPVRCSNCGRSCPKDKAIKRF 55
R PTK++ R K +G GH P+ C+NC PKDKAIK+F
Sbjct 50 RKPTKQQGINRVKGKTTSINGCGHTQPIHCTNCAPWVPKDKAIKKF 95
> Hs17474677
Length=129
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/63 (44%), Positives = 41/63 (65%), Gaps = 4/63 (6%)
Query 48 KDKAIKRFSVRNIVDASSQRDLREASAYHTYNLPKLYIK-HCYCISCAIHSRVVRVRSRE 106
+DKAIK F ++N V+A + R + E S ++T +LY K HC +SCAIH +VVR S E
Sbjct 25 QDKAIK-FVIKNKVEAIAIRAISEVSVFNTCA-SQLYEKLHCD-MSCAIHGKVVRSHSHE 81
Query 107 RRR 109
++
Sbjct 82 AQK 84
> Hs18597386
Length=95
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 17 MPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQR 67
M +RRN K H+ P+ C N KDKAIK+ ++N + ++ R
Sbjct 1 MTMRRRNNSHGKKDHSHIQPIYCMNLALCVAKDKAIKKLIIQNTAETAAIR 51
> Hs22056634
Length=255
Score = 34.3 bits (77), Expect = 0.053, Method: Compositional matrix adjust.
Identities = 13/28 (46%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 30 GRGHVNPVRCSNCGRSCPKDKAIKRFSV 57
G GHV + C NC + P +KAIK+F +
Sbjct 228 GHGHVLSIHCMNCAQCVPDNKAIKKFII 255
> Hs18600782
Length=453
Score = 30.8 bits (68), Expect = 0.69, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 27/51 (52%), Gaps = 3/51 (5%)
Query 7 SGLRRQEAERMPTK-RRNAGRSKHGRGH--VNPVRCSNCGRSCPKDKAIKR 54
SGLR P RR A S+H RGH P+RC+ C R+ P+ ++R
Sbjct 67 SGLRPHACPLCPKAFRRPAHLSRHLRGHGPQPPLRCAACPRTFPEPAQLRR 117
> At4g00990
Length=884
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 86 KHCYCISCAIHSRVVRVRSRERRRVRTNPQAR 117
K CYC C SRV+ + + +R RT+ + R
Sbjct 100 KKCYCFDCIKRSRVLTILNLDRYSERTHEEVR 131
> At1g77460
Length=2110
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 24/81 (29%), Positives = 36/81 (44%), Gaps = 6/81 (7%)
Query 6 VSGLRRQEAERMPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASS 65
VS R A+ P +RNA R+ H PV C K A RF++ ++VD+
Sbjct 738 VSAFTRILADGSPEGKRNASRALHQLLKNFPV----C--DVLKGSAQCRFAILSLVDSLK 791
Query 66 QRDLREASAYHTYNLPKLYIK 86
D+ A A++ + L K
Sbjct 792 SIDVDSADAFNILEVVALLAK 812
> CE28395
Length=624
Score = 28.1 bits (61), Expect = 4.0, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 26/52 (50%), Gaps = 3/52 (5%)
Query 30 GRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYHTYNLP 81
G+ H+NP + G+ D+ V+N++DAS QR ++ Y +P
Sbjct 574 GQRHLNPKFQVHAGKV---DQLEANDKVKNVLDASPQRTWKDIDGYGVLGVP 622
> Hs10835041
Length=648
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 13/25 (52%), Positives = 17/25 (68%), Gaps = 2/25 (8%)
Query 23 NAGRSKHGRGH--VNPVRCSNCGRS 45
+A +KH RGH V P RC+ CG+S
Sbjct 577 SAAFAKHLRGHASVRPCRCNECGKS 601
> Hs7657665
Length=487
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 2/32 (6%)
Query 18 PTKRRN-AGRSKHGRGH-VNPVRCSNCGRSCP 47
P + R+ + +KH R H P RCS CG CP
Sbjct 407 PQRSRDFSAMTKHLRTHGAAPYRCSLCGAGCP 438
> CE00378
Length=3343
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 2 FLIGVSGLRRQEAERMPTKRRNAGRSKHGRGHVNPVRCSNCGRSCPK 48
F + + +RR+ ++ P ++ RS GHV P R + RS K
Sbjct 3243 FSVCLLAIRRRWRQKSPGDQKQTERSNGWTGHVMPRRRGHINRSMVK 3289
> Hs20472542
Length=402
Score = 27.3 bits (59), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 22/36 (61%), Gaps = 2/36 (5%)
Query 21 RRNAGRSKHGRGHV--NPVRCSNCGRSCPKDKAIKR 54
R ++G +H R H P +C+ CG++ P+ A+K+
Sbjct 353 RHSSGLVEHQRLHTGEKPYKCNECGKAFPRSSALKQ 388
> At4g01920
Length=658
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 23/100 (23%), Positives = 37/100 (37%), Gaps = 22/100 (22%)
Query 9 LRRQEAERMPTKRRNAGRSKHGRGHVNPVR----CSNCGRSCP-----------KDKAIK 53
+R+ A+ ++ KH + P R C+ CGR CP D
Sbjct 50 VRKNCADEPSEYIKHPSHPKHTLQLLRPERPTNFCNLCGRMCPIFYRCDLCDFDMDLYCS 109
Query 54 RFSVRNIVDASSQRDLREASAYHTYNLPKLYIKHCYCISC 93
++ ++D S + +H L K +IK C C C
Sbjct 110 KYPPPEVIDISK-------THHHRLTLLKKWIKQCICAKC 142
> CE28394
Length=678
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query 30 GRGHVNPVRCSNCGRSCPKDKAIKRFSVRNIVDASSQRDLREASAYHTYNLPKLYIKHCY 89
G+ H+NP + G+ D+ V+N++DAS QR ++ Y LP H Y
Sbjct 574 GQRHLNPKFQVHAGKV---DQLEANDKVKNVLDASPQRTWKDIDGYD--GLPDSGRSHYY 628
Query 90 CI 91
I
Sbjct 629 FI 630
> Hs4758228
Length=784
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 5/44 (11%)
Query 12 QEAERMPTKRRNAGRSKHGRGHVNP--VRCSNCGRSCPKDKAIK 53
Q +E PT A H RGH P C+ CG++ PK +K
Sbjct 439 QCSETFPTA---ATLEAHKRGHTGPRPFACAQCGKAFPKAYLLK 479
> Hs17477058
Length=279
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 20 KRRNAGRSKHGRGHVNPVRCSNCGRSCPKDKAIK 53
KRRN +K G ++ +R +NC K+KAIK
Sbjct 5 KRRNNSHAKKGHSYMQLIRSTNCVPCGSKNKAIK 38
Lambda K H
0.323 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1213511838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40