bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
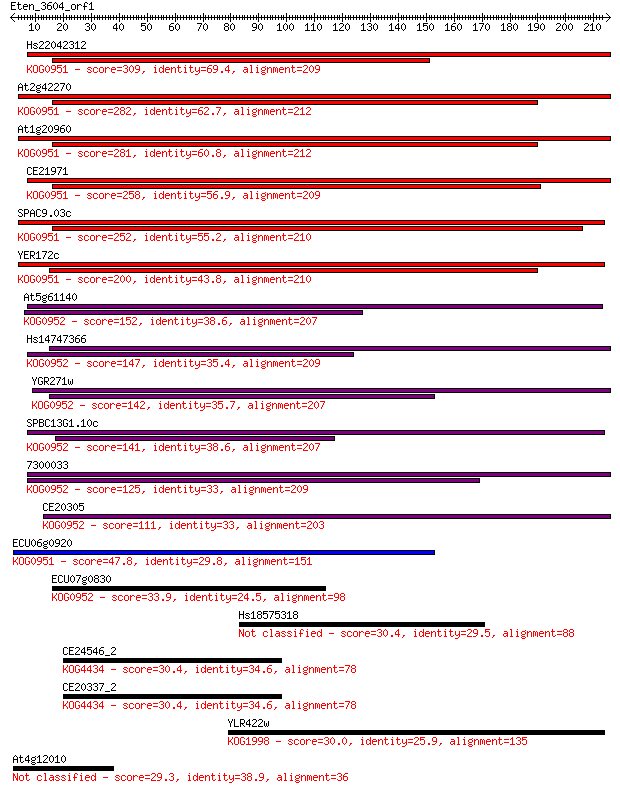
Query= Eten_3604_orf1
Length=215
Score E
Sequences producing significant alignments: (Bits) Value
Hs22042312 309 3e-84
At2g42270 282 4e-76
At1g20960 281 5e-76
CE21971 258 9e-69
SPAC9.03c 252 5e-67
YER172c 200 2e-51
At5g61140 152 6e-37
Hs14747366 147 2e-35
YGR271w 142 6e-34
SPBC13G1.10c 141 1e-33
7300033 125 7e-29
CE20305 111 1e-24
ECU06g0920 47.8 2e-05
ECU07g0830 33.9 0.27
Hs18575318 30.4 2.9
CE24546_2 30.4 2.9
CE20337_2 30.4 2.9
YLR422w 30.0 3.8
At4g12010 29.3 5.9
> Hs22042312
Length=2136
Score = 309 bits (791), Expect = 3e-84, Method: Compositional matrix adjust.
Identities = 145/209 (69%), Positives = 176/209 (84%), Gaps = 0/209 (0%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQ 66
LKPTLS+IEL R+FSLSSEFK + VREEEKLELQ+L+ERVPIPVK S +EPS+K+NVLLQ
Sbjct 1005 LKPTLSEIELFRVFSLSSEFKNITVREEEKLELQKLLERVPIPVKESIEEPSAKINVLLQ 1064
Query 67 AYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWS 126
A+IS+LKLEGFA+MADMVYV QSA R+MRAIF+I L RGWAQL + L LC + RMW
Sbjct 1065 AFISQLKLEGFALMADMVYVTQSAGRLMRAIFEIVLNRGWAQLTDKTLNLCKMIDKRMWQ 1124
Query 127 TMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKL 186
+M PLRQF+ LPEE+++KIEKK+ PF+R YDL EIGEL+R+PKMGK +H+ +H FPKL
Sbjct 1125 SMCPLRQFRKLPEEVVKKIEKKNFPFERLYDLNHNEIGELIRMPKMGKTIHKYVHLFPKL 1184
Query 187 ELAAFVQPLSRSCLVVELTITPDFQWDPK 215
EL+ +QP++RS L VELTITPDFQWD K
Sbjct 1185 ELSVHLQPITRSTLKVELTITPDFQWDEK 1213
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 37/136 (27%), Positives = 74/136 (54%), Gaps = 2/136 (1%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKL 74
L+ + S ++E++ +P+R E L++L ++VP + ++P K N+LLQA++S+++L
Sbjct 1845 LIEIISNAAEYENIPIRHHEDNLLRQLAQKVPHKLNNPKFNDPHVKTNLLLQAHLSRMQL 1904
Query 75 EGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQF 134
+ +D + A R+++A D+ GW A+ A++L V MWS + L+Q
Sbjct 1905 SA-ELQSDTEEILSKAIRLIQACVDVLSSNGWLSPALAAMELAQMVTQAMWSKDSYLKQL 1963
Query 135 KVLPEELLRKIEKKDL 150
E +++ K +
Sbjct 1964 PHFTSEHIKRCTDKGV 1979
> At2g42270
Length=2172
Score = 282 bits (721), Expect = 4e-76, Method: Compositional matrix adjust.
Identities = 133/212 (62%), Positives = 172/212 (81%), Gaps = 0/212 (0%)
Query 4 RRHLKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNV 63
+LKPT++DIEL RLFSLS EFKY+ VR++EK+EL +L++RVPIPVK + ++PS+K+NV
Sbjct 1027 NENLKPTMNDIELCRLFSLSEEFKYVTVRQDEKMELAKLLDRVPIPVKETLEDPSAKINV 1086
Query 64 LLQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGR 123
LLQ YISKLKLEG ++ +DMVY+ QSA R++RAIF+I L+RGWAQL+ +AL L V R
Sbjct 1087 LLQVYISKLKLEGLSLTSDMVYITQSAGRLLRAIFEIVLKRGWAQLSQKALNLSKMVGKR 1146
Query 124 MWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSF 183
MWS TPL QF +P+E+L K+EK DL ++RYYDL+S E+GEL+ PKMG+ LH+ IH F
Sbjct 1147 MWSVQTPLWQFPGIPKEILMKLEKNDLVWERYYDLSSQELGELICNPKMGRPLHKYIHQF 1206
Query 184 PKLELAAFVQPLSRSCLVVELTITPDFQWDPK 215
PKL+LAA VQP+SRS L VELT+TPDF WD K
Sbjct 1207 PKLKLAAHVQPISRSVLQVELTVTPDFHWDDK 1238
Score = 55.8 bits (133), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 45/180 (25%), Positives = 86/180 (47%), Gaps = 7/180 (3%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKG-SPDEPSSKVNVLLQAYISKLKL 74
LL + + +SE+ +P+R E+ ++RL+ + +P K + LLQA+ S+ K+
Sbjct 1873 LLEILTSASEYDLIPIRPGEEDAVRRLINHQRFSFQNPRCTDPRVKTSALLQAHFSRQKI 1932
Query 75 EGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQF 134
G +M D V SA+R+++A+ D+ G LA+ A+++ V MW + L Q
Sbjct 1933 SGNLVM-DQCEVLLSATRLLQAMVDVISSNGCLNLALLAMEVSQMVTQGMWDRDSMLLQL 1991
Query 135 KVLPEELLRKI-EKKDLPFDRYYDLTSTEIGELVRVPKMGKL----LHRLIHSFPKLELA 189
++L ++ E + +DL E + + +M + R + FP ++L
Sbjct 1992 PHFTKDLAKRCHENPGNNIETIFDLVEMEDDKRQELLQMSDAQLLDIARFCNRFPNIDLT 2051
> At1g20960
Length=2171
Score = 281 bits (720), Expect = 5e-76, Method: Compositional matrix adjust.
Identities = 129/212 (60%), Positives = 172/212 (81%), Gaps = 0/212 (0%)
Query 4 RRHLKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNV 63
HLKPT+ DI+L RLFSLS EFKY+ VR++EK+EL +L++RVPIP+K + +EPS+K+NV
Sbjct 1026 NEHLKPTMGDIDLYRLFSLSDEFKYVTVRQDEKMELAKLLDRVPIPIKETLEEPSAKINV 1085
Query 64 LLQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGR 123
LLQAYIS+LKLEG ++ +DMVY+ QSA R++RA+++I L+RGWAQLA +AL L V R
Sbjct 1086 LLQAYISQLKLEGLSLTSDMVYITQSAGRLVRALYEIVLKRGWAQLAEKALNLSKMVGKR 1145
Query 124 MWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSF 183
MWS TPLRQF L ++L ++EKKDL ++RYYDL++ E+GEL+R PKMGK LH+ IH F
Sbjct 1146 MWSVQTPLRQFHGLSNDILMQLEKKDLVWERYYDLSAQELGELIRSPKMGKPLHKFIHQF 1205
Query 184 PKLELAAFVQPLSRSCLVVELTITPDFQWDPK 215
PK+ L+A VQP++R+ L VELT+TPDF WD K
Sbjct 1206 PKVTLSAHVQPITRTVLNVELTVTPDFLWDEK 1237
Score = 62.4 bits (150), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 48/180 (26%), Positives = 85/180 (47%), Gaps = 7/180 (3%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKG-SPDEPSSKVNVLLQAYISKLKL 74
LL + + +SE+ +P+R E+ ++RL+ + +P K N LLQA+ S+ +
Sbjct 1872 LLEILTSASEYDMIPIRPGEEDTVRRLINHQRFSFENPKCTDPHVKANALLQAHFSRQNI 1931
Query 75 EGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQF 134
G M D V SA+R+++A+ D+ GW LA+ A+++ V MW + L Q
Sbjct 1932 GGNLAM-DQRDVLLSATRLLQAMVDVISSNGWLNLALLAMEVSQMVTQGMWERDSMLLQL 1990
Query 135 KVLPEELLRKI-EKKDLPFDRYYDLTSTEIGELVRVPKMGKL----LHRLIHSFPKLELA 189
++L ++ E + +DL E E + KM + R + FP ++L
Sbjct 1991 PHFTKDLAKRCQENPGKNIETVFDLVEMEDEERQELLKMSDAQLLDIARFCNRFPNIDLT 2050
> CE21971
Length=2145
Score = 258 bits (658), Expect = 9e-69, Method: Compositional matrix adjust.
Identities = 119/209 (56%), Positives = 158/209 (75%), Gaps = 0/209 (0%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQ 66
L T SDI+L R+FS+SSEFK + VR+EEKLELQ++ E PIP+K + DE S+K NVLLQ
Sbjct 998 LVETCSDIDLFRIFSMSSEFKLLSVRDEEKLELQKMAEHAPIPIKENLDEASAKTNVLLQ 1057
Query 67 AYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWS 126
AYIS+LKLEGFA+ ADMV+V QSA R+ RA+F+I L RGWA LA + L LC V R W
Sbjct 1058 AYISQLKLEGFALQADMVFVAQSAGRLFRALFEIVLWRGWAGLAQKVLTLCKMVTQRQWG 1117
Query 127 TMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKL 186
++ PL QFK +P E++R I+KK+ FDR YDL ++G+L+++PKMGK L + I FPKL
Sbjct 1118 SLNPLHQFKKIPSEVVRSIDKKNYSFDRLYDLDQHQLGDLIKMPKMGKPLFKFIRQFPKL 1177
Query 187 ELAAFVQPLSRSCLVVELTITPDFQWDPK 215
E+ +QP++R+ + +ELTITPDF+WD K
Sbjct 1178 EMTTLIQPITRTTMRIELTITPDFKWDEK 1206
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 53/185 (28%), Positives = 89/185 (48%), Gaps = 18/185 (9%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSP-DEPSSKVNVLLQAYISKLKL 74
L+ + S SSEF +P+R +E + L++L ER+P +K +P KVN+L+ A++S++KL
Sbjct 1844 LIEIISASSEFGNVPMRHKEDVILRQLAERLPGQLKNQKFTDPHVKVNLLIHAHLSRVKL 1903
Query 75 EGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQF 134
+ D + A R+++A D+ GW AI A++L + M+S L+Q
Sbjct 1904 TA-ELNKDTELIVLRACRLVQACVDVLSSNGWLSPAIHAMELSQMLTQAMYSNEPYLKQL 1962
Query 135 KVLPEELLRKIEKK---------DLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPK 185
LL + + K +L D D+ E EL V R + +P
Sbjct 1963 PHCSAALLERAKAKEVTSVFELLELENDDRSDILQMEGAELADVA-------RFCNHYPS 2015
Query 186 LELAA 190
+E+A
Sbjct 2016 IEVAT 2020
> SPAC9.03c
Length=2176
Score = 252 bits (643), Expect = 5e-67, Method: Compositional matrix adjust.
Identities = 116/210 (55%), Positives = 159/210 (75%), Gaps = 0/210 (0%)
Query 4 RRHLKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNV 63
R L T S IEL R+FS S EFK++PVREEEK+EL +L+ERVPIP++ DEP++K+N
Sbjct 1040 NRLLMQTTSFIELFRVFSFSDEFKHIPVREEEKVELAKLLERVPIPIRERLDEPAAKINA 1099
Query 64 LLQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGR 123
LLQ+YIS+ +L+GFA++ADMVYV QSA RIMRAIF+I LRRGW+ +A +L C ++ R
Sbjct 1100 LLQSYISRQRLDGFALVADMVYVTQSAGRIMRAIFEISLRRGWSSVATLSLDTCKMIEKR 1159
Query 124 MWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSF 183
+W TM+PLRQF P E++R++EKK+ P+ RY+DL E+GELV VPK G+ ++ ++ SF
Sbjct 1160 LWPTMSPLRQFPNCPSEVIRRVEKKEFPWQRYFDLDPAELGELVGVPKEGRRVYNMVQSF 1219
Query 184 PKLELAAFVQPLSRSCLVVELTITPDFQWD 213
P+L + A VQP++RS + VEL I F WD
Sbjct 1220 PRLSVEAHVQPITRSLVRVELVINSQFNWD 1249
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 45/200 (22%), Positives = 96/200 (48%), Gaps = 12/200 (6%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKG-SPDEPSSKVNVLLQAYISKLKL 74
LL + + ++E++ +P+R+ E + L+R+ R+P+ + + ++P +K +LL A+ S+ +L
Sbjct 1881 LLEIVTSAAEYEQLPIRKYEDIVLERIHSRLPVRLSNPNYEDPHTKSFILLAAHFSRFEL 1940
Query 75 EGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQF 134
++ D ++ ++ A D G IR +++ V +W +PL+Q
Sbjct 1941 PP-GLVIDQKFILTRVHNLLGACVDTLSSEGHLIACIRPMEMSQMVTQALWDRDSPLKQI 1999
Query 135 KVLPEELLRKIEKKDLP--FDRYYDLTSTEIGELVRVPKMG-KLLHRLIHSFPKLEL--- 188
+ L+ + K+ + FD DL + EL+ + I+ +P +++
Sbjct 2000 PYFDDALIERCNKEGVHDVFD-IIDLDDEKRTELLHMDNAHLAKCAEFINKYPDIDIDFE 2058
Query 189 ---AAFVQPLSRSCLVVELT 205
+ V S S L+V+LT
Sbjct 2059 IEDSEDVHANSPSVLIVQLT 2078
> YER172c
Length=2163
Score = 200 bits (508), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 92/211 (43%), Positives = 138/211 (65%), Gaps = 1/211 (0%)
Query 4 RRHLKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNV 63
R L + I+L R+FS+S EFKY+ VR EEK EL++L+E+ PIP++ D+P +KVNV
Sbjct 1019 NRELDEHTTQIDLFRIFSMSEEFKYVSVRYEEKRELKQLLEKAPIPIREDIDDPLAKVNV 1078
Query 64 LLQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGR 123
LLQ+Y S+LK EGFA+ +D+V++ Q+A R++RA+F+ICL+RGW L LC +
Sbjct 1079 LLQSYFSQLKFEGFALNSDIVFIHQNAGRLLRAMFEICLKRGWGHPTRMLLNLCKSATTK 1138
Query 124 MWSTMTPLRQFKVLPEELLRKIEKKDLPFDRYYDL-TSTEIGELVRVPKMGKLLHRLIHS 182
MW T PLRQFK P E+++++E +P+ Y L T E+G +R K GK ++ L+
Sbjct 1139 MWPTNCPLRQFKTCPVEVIKRLEASTVPWGDYLQLETPAEVGRAIRSEKYGKQVYDLLKR 1198
Query 183 FPKLELAAFVQPLSRSCLVVELTITPDFQWD 213
FPK+ + QP++RS + + I D+ WD
Sbjct 1199 FPKMSVTCNAQPITRSVMRFNIEIIADWIWD 1229
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 46/181 (25%), Positives = 85/181 (46%), Gaps = 10/181 (5%)
Query 15 ELLRLFSLSSEFKYMPVREEEKLELQRLMERVPI--PVKGSPDEPSSKVNVLLQAYISKL 72
+L + S + EF+ +P+R+ ++ L +L +R+P+ P S S KV +LLQAY S+L
Sbjct 1878 NMLYVLSTAVEFESVPLRKGDRALLVKLSKRLPLRFPEHTSSGSVSFKVFLLLQAYFSRL 1937
Query 73 KLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLR 132
+L D+ + + ++ + DI G+ A A+ L + +W PLR
Sbjct 1938 ELP-VDFQNDLKDILEKVVPLINVVVDILSANGYLN-ATTAMDLAQMLIQGVWDVDNPLR 1995
Query 133 QFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGE----LVRVPKMGKLLHRLIHSFPKLEL 188
Q ++L K K++ + YD+ + E E L + ++++P +EL
Sbjct 1996 QIPHFNNKILEKC--KEINVETVYDIMALEDEERDEILTLTDSQLAQVAAFVNNYPNVEL 2053
Query 189 A 189
Sbjct 2054 T 2054
> At5g61140
Length=2137
Score = 152 bits (383), Expect = 6e-37, Method: Compositional matrix adjust.
Identities = 80/213 (37%), Positives = 127/213 (59%), Gaps = 23/213 (10%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERV-PIPVKGSPDEPSSKVNVLL 65
LK +++ E++ + + SSEF+ + VREEE+ EL+ L P+ VKG P K+++L+
Sbjct 1032 LKRHMNESEIINMVAHSSEFENIVVREEEQHELETLARSCCPLEVKGGPSNKHGKISILI 1091
Query 66 QAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMW 125
Q YIS+ ++ F++++D Y+ S +RIMRA+F+ICLR+GW ++ + L+ C V ++W
Sbjct 1092 QLYISRGSIDAFSLVSDASYISASLARIMRALFEICLRKGWCEMTLFMLEYCKAVDRQLW 1151
Query 126 STMTPLRQFKVLPEELLRKIEKKDLPFDR------YYDLTSTEIGELVRVPKMGKLLHRL 179
PLRQF ++DLP DR Y++ EIG L+R G R
Sbjct 1152 PHQHPLRQF------------ERDLPSDRRDDLDHLYEMEEKEIGALIRYNPGG----RH 1195
Query 180 IHSFPKLELAAFVQPLSRSCLVVELTITPDFQW 212
+ FP ++LAA V P++R+ L V+L ITP+F W
Sbjct 1196 LGYFPSIQLAATVSPITRTVLKVDLLITPNFIW 1228
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 49/122 (40%), Gaps = 30/122 (24%)
Query 6 HLKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSP-DEPSSKVNVL 64
++ P S L + + +SE+ +PVR E+ + L +RV PV + D+P K N+L
Sbjct 1880 NIGPDTSLEAFLHILAGASEYDELPVRHNEENYNKTLSDRVRYPVDNNHLDDPHVKANLL 1939
Query 65 LQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRM 124
QA I DIC GW ++ ++L V M
Sbjct 1940 FQAMI-----------------------------DICANSGWLSSSLTCMRLLQMVMQGM 1970
Query 125 WS 126
WS
Sbjct 1971 WS 1972
> Hs14747366
Length=1866
Score = 147 bits (370), Expect = 2e-35, Method: Compositional matrix adjust.
Identities = 70/202 (34%), Positives = 120/202 (59%), Gaps = 1/202 (0%)
Query 15 ELLRLFSLSSEFKYMPVREEEKLELQRLMER-VPIPVKGSPDEPSSKVNVLLQAYISKLK 73
++ + S + EF + VREEE EL L+ + G + K+N+LLQ YIS+ +
Sbjct 674 DIFAIVSKAEEFDQIKVREEEIEELDTLLSNFCELSTPGGVENSYGKINILLQTYISRGE 733
Query 74 LEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQ 133
++ F++++D YV Q+A+RI+RA+F+I LR+ W + R L L + R+W +PLRQ
Sbjct 734 MDSFSLISDSAYVAQNAARIVRALFEIALRKRWPTMTYRLLNLSKVIDKRLWGWASPLRQ 793
Query 134 FKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQ 193
F +LP +L ++E+K L D+ D+ EIG ++ +G + + +H P + + A +Q
Sbjct 794 FSILPPHILTRLEEKKLTVDKLKDMRKDEIGHILHHVNIGLKVKQCVHQIPSVMMEASIQ 853
Query 194 PLSRSCLVVELTITPDFQWDPK 215
P++R+ L V L+I DF W+ +
Sbjct 854 PITRTVLRVTLSIYADFTWNDQ 875
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 39/119 (32%), Positives = 57/119 (47%), Gaps = 2/119 (1%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVK-GSPDEPSSKVNVLL 65
LKP S ELL + S + E+ +PVR E L + +PI S D P +K ++LL
Sbjct 1500 LKPECSTEELLSILSDAEEYTDLPVRHNEDHMNSELAKCLPIESNPHSFDSPHTKAHLLL 1559
Query 66 QAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEV-QGR 123
QA++S+ L D V A R+ +A+ D+ +GW + L V QGR
Sbjct 1560 QAHLSRAMLPCPDYDTDTKTVLDQALRVCQAMLDVAANQGWLVTVLNITNLIQMVIQGR 1618
> YGR271w
Length=1967
Score = 142 bits (357), Expect = 6e-34, Method: Compositional matrix adjust.
Identities = 74/208 (35%), Positives = 121/208 (58%), Gaps = 2/208 (0%)
Query 9 PTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLM-ERVPIPVKGSPDEPSSKVNVLLQA 67
P ++ ++L + S+SSEF + REEE EL+RL E V + D P K NVLLQA
Sbjct 821 PRATEADVLSMISMSSEFDGIKFREEESKELKRLSDESVECQIGSQLDTPQGKANVLLQA 880
Query 68 YISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWST 127
YIS+ ++ A+ +D YV Q++ RI RA+F I + R W + + L +C ++ R+W+
Sbjct 881 YISQTRIFDSALSSDSNYVAQNSVRICRALFLIGVNRRWGKFSNVMLNICKSIEKRLWAF 940
Query 128 MTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLE 187
PL QF LPE ++R+I + +L + E+GELV K G L++++ FPK+
Sbjct 941 DHPLCQFD-LPENIIRRIRDTKPSMEHLLELEADELGELVHNKKAGSRLYKILSRFPKIN 999
Query 188 LAAFVQPLSRSCLVVELTITPDFQWDPK 215
+ A + P++ + + + + + PDF WD +
Sbjct 1000 IEAEIFPITTNVMRIHIALGPDFVWDSR 1027
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 40/146 (27%), Positives = 70/146 (47%), Gaps = 13/146 (8%)
Query 15 ELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGS-PDE-------PSSKVNVLLQ 66
E+LR SL+ E+ +PVR E + + + ++ V+ + DE P K +LLQ
Sbjct 1658 EVLRWLSLAVEYNELPVRGGEIIMNEEMSQQSRYSVESTFTDEFELPMWDPHVKTFLLLQ 1717
Query 67 AYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWS 126
A++S++ L + D V V + RI++A D+ G+ + +++ ++ W
Sbjct 1718 AHLSRVDLPIADYIQDTVSVLDQSLRILQAYIDVASELGYFHTVLTMIKMMQCIKQGYWY 1777
Query 127 TMTPLRQFKVLPEELLRKIEKKDLPF 152
P+ VLP LR+I KD F
Sbjct 1778 EDDPV---SVLPGLQLRRI--KDYTF 1798
> SPBC13G1.10c
Length=1935
Score = 141 bits (355), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 80/210 (38%), Positives = 119/210 (56%), Gaps = 5/210 (2%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERV-PIPVKGSPDEPSSKVNVLL 65
LK +S+ +++ L S SEF + RE E EL+ LME P ++ S S KVNV+L
Sbjct 811 LKSKMSEADIIALLSQCSEFSQIKSRENEHRELESLMENSSPCQLRDSISNTSGKVNVIL 870
Query 66 QAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMW 125
Q+YIS+ +E F + +D YV Q+A RI RA+F+I + R WA A L L + R W
Sbjct 871 QSYISRAHVEDFTLTSDTNYVAQNAGRITRALFEIAMSRTWAS-AFTILSLNKSIDRRQW 929
Query 126 STMTPLRQFKVLPEELLRKIEKK--DLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSF 183
S PL QF LP +L K+E + L + D+++ E+G+L+ KMG + + I
Sbjct 930 SFEHPLLQFD-LPHDLAVKVENQCGSLSLEELSDMSTGELGDLIHNRKMGPTVKKFISKL 988
Query 184 PKLELAAFVQPLSRSCLVVELTITPDFQWD 213
P L + + PL+++ L + L ITP+F WD
Sbjct 989 PLLNINVDLLPLTKNVLRLVLNITPNFNWD 1018
Score = 33.5 bits (75), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 23/103 (22%), Positives = 50/103 (48%), Gaps = 3/103 (2%)
Query 17 LRLFSLSSEFKYMPVREEEKL---ELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLK 73
L+L + +SEF + +R E L E+ + ++ + + K +L QA++++LK
Sbjct 1652 LQLLAEASEFDDLAIRHNEDLINIEINKSLKYSAACLNLPMVDAHVKAFILTQAHMARLK 1711
Query 74 LEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQL 116
L + D V RI+++ D+ G++ + ++ + L
Sbjct 1712 LPVDDYVTDTSTVLDQVIRIIQSYIDVSAELGYSHVCLQYISL 1754
> 7300033
Length=1809
Score = 125 bits (314), Expect = 7e-29, Method: Compositional matrix adjust.
Identities = 69/210 (32%), Positives = 114/210 (54%), Gaps = 1/210 (0%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMER-VPIPVKGSPDEPSSKVNVLL 65
+KP ++ E+L + S + EF+ + VR++E EL L I G + KVN+L+
Sbjct 638 MKPFMTQAEILAMISQAQEFQQLKVRDDEMEELDELKNAYCKIKPYGGSENVHGKVNILI 697
Query 66 QAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMW 125
Q Y+S ++ F++ +DM Y+ + RI RA+F I LR+ A L+ LQLC + R W
Sbjct 698 QTYLSNGYVKSFSLSSDMSYITTNIGRISRALFSIVLRQNNAVLSGNMLQLCKMFERRQW 757
Query 126 STMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPK 185
L+QF + E + K+E++ L R D+ E+ E +R L+ R H P
Sbjct 758 DFDCHLKQFPTINAETIDKLERRGLSVYRLRDMEHRELKEWLRSNTYADLVIRSAHELPL 817
Query 186 LELAAFVQPLSRSCLVVELTITPDFQWDPK 215
LE+ A +QP++R+ L +++ I P F W+ +
Sbjct 818 LEVEASLQPITRTVLRIKVDIWPSFTWNDR 847
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/163 (25%), Positives = 70/163 (42%), Gaps = 3/163 (1%)
Query 7 LKPTLSDIELLRLFSLSSEFKYMPVREEEKLELQRLMERVPI-PVKGSPDEPSSKVNVLL 65
L+P ++ ++L + S EF +PVR E + + E P S D +K +LL
Sbjct 1471 LQPGMNTKKVLLAIADSYEFDQLPVRHNEDKHNEEMAEVSRFRPPSSSWDSSYTKTFLLL 1530
Query 66 QAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMW 125
QA+ ++ L + D +A+R+M+A+ D RGW + QL V W
Sbjct 1531 QAHFARQSLPNSDYLTDTKSALDNATRVMQAMVDYTAERGWLSTTLVVQQLMQSVIQARW 1590
Query 126 STMTPLRQFKVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVR 168
+ + E+ L ++P D Y LT + EL +
Sbjct 1591 FDGSEFLTLPGVNEDNLDAF--LNIPHDDYDYLTLPVLKELCK 1631
> CE20305
Length=1798
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 67/208 (32%), Positives = 117/208 (56%), Gaps = 7/208 (3%)
Query 13 DIELLRLFSLSSEFKYMPVREEEKLELQRLME-RVPIPVKGSP-DEPSSKVNVLLQAYIS 70
D ++ L S+S+EF + REEE +L+ LM + V+G + KVNVLLQ+ IS
Sbjct 638 DDMVIGLISMSTEFANLKCREEEIGDLEELMSYGCMMNVRGGGLASVAGKVNVLLQSLIS 697
Query 71 KLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTP 130
+ A+M++ +YVQQ+A R+ RA+F++ L+ GW+Q A L + V+ +MW
Sbjct 698 RTSTRNSALMSEQLYVQQNAGRLCRAMFEMVLKNGWSQAANAFLGIAKCVEKQMWMNQCA 757
Query 131 LRQF---KVLPEELLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLE 187
LRQF +P + KIE+K +L++ ++G + G L+ + P+++
Sbjct 758 LRQFIQIINIPITWIEKIERKKARESDLLELSAKDLGYMFSCD--GDRLYTYLRYLPRMD 815
Query 188 LAAFVQPLSRSCLVVELTITPDFQWDPK 215
+ A +P++ + + VE+T+TP F W+ +
Sbjct 816 VQAKFKPITYTIVQVEVTLTPTFIWNDQ 843
> ECU06g0920
Length=1481
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 45/157 (28%), Positives = 76/157 (48%), Gaps = 28/157 (17%)
Query 2 RPRRHLKPTLSDI----ELLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEP 57
R R L LS + + ++ + EF + + E+ L+ L VPIP +
Sbjct 695 RDARRLFDGLSHVMLEPSIFQILEKAREFSDLSIDEKAMESLRGL---VPIPTE------ 745
Query 58 SSKVNVLLQAYISKLKLEGFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQL- 116
S VLLQ Y++ ++E ++ Q+ R+ RA+F+I +R+ +L I + L
Sbjct 746 -SSFGVLLQCYVAN-RIESTSL-------SQNLCRMFRALFEIGVRK---RLGISKMILG 793
Query 117 -CNEVQGRMWSTMTPLRQFKVLPEELLRKIEKKDLPF 152
C + R++ TPLRQF + LR +E K++PF
Sbjct 794 WCKAAEHRIFPYQTPLRQF-ADDKNALRDLEMKEIPF 829
> ECU07g0830
Length=1058
Score = 33.9 bits (76), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 24/98 (24%), Positives = 51/98 (52%), Gaps = 3/98 (3%)
Query 16 LLRLFSLSSEFKYMPVREEEKLELQRLMERVPIPVKGSPDEPSSKVNVLLQAYISKLKLE 75
+LRL + E + R+E++ ++ L E + I + S + K+ L++A+I + +
Sbjct 605 ILRLCFEARELSALRCRDEDEESIKELCEDLGIKYEVSVE---CKLMALVKAHIKRHPVT 661
Query 76 GFAMMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRA 113
F++M D YV ++ RI+ + + + +G L +R
Sbjct 662 RFSLMCDGEYVIKNLRRILMGLCQVLMFQGTHLLVVRC 699
> Hs18575318
Length=139
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 26/88 (29%), Positives = 39/88 (44%), Gaps = 2/88 (2%)
Query 83 MVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCNEVQGRMWSTMTPLRQFKVLPEELL 142
+ VQ + +IM + R+ W + L + EV G W + Q KVL E+ L
Sbjct 9 LTIVQATNEKIMYLATFLTARKNWNTERLSQLGITGEV-GAGW-ILRDQVQVKVLREKRL 66
Query 143 RKIEKKDLPFDRYYDLTSTEIGELVRVP 170
RK D F + L++T + L VP
Sbjct 67 RKQRTVDELFQSWLALSATSVSLLDAVP 94
> CE24546_2
Length=535
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 44/86 (51%), Gaps = 11/86 (12%)
Query 20 FSLSSEF-KYMPVREEEKLELQRLMERVPIPVKGSPDEPSS-----KVNVLLQAYISKLK 73
F S ++ K + RE + E+ RLM+++ ++P S K VLL AY+S+L
Sbjct 40 FEFSKQYNKEIQERETDDYEVPRLMKQIAGVNDKGKEQPLSQPYALKSRVLLHAYLSRLP 99
Query 74 LE--GFAMMADMVYVQQSASRIMRAI 97
LE A+ D Y+ +RI+R +
Sbjct 100 LESQNDALEEDQSYI---ITRILRFV 122
> CE20337_2
Length=362
Score = 30.4 bits (67), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 44/86 (51%), Gaps = 11/86 (12%)
Query 20 FSLSSEF-KYMPVREEEKLELQRLMERVPIPVKGSPDEPSS-----KVNVLLQAYISKLK 73
F S ++ K + RE + E+ RLM+++ ++P S K VLL AY+S+L
Sbjct 40 FEFSKQYNKEIQERETDDYEVPRLMKQIAGVNDKGKEQPLSQPYALKSRVLLHAYLSRLP 99
Query 74 LE--GFAMMADMVYVQQSASRIMRAI 97
LE A+ D Y+ +RI+R +
Sbjct 100 LESQNDALEEDQSYI---ITRILRFV 122
> YLR422w
Length=1932
Score = 30.0 bits (66), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 35/140 (25%), Positives = 65/140 (46%), Gaps = 9/140 (6%)
Query 79 MMADMVYVQQSASRIMRAIFDICLRRGWAQLAIRALQLCN--EVQGRMWSTMTPLRQFKV 136
+MAD + + +S ++ + + C + AI + C E+ +M+S +R+ +
Sbjct 885 IMADQILLIESIPSMLETMTNHCKVEDLVRFAIGLFECCQEKEMNQKMYSRPLSVREEEY 944
Query 137 LPEE---LLRKIEKKDLPFDRYYDLTSTEIGELVRVPKMGKLLHRLIHSFPKLELAAFVQ 193
L + LL+ I KK L Y LT+TE + +R+ + K L L+ + + F
Sbjct 945 LNTKFNCLLKLINKKVL--QNY--LTNTESVDKLRLQFLSKTLEWLLTPYTPGDDKCFHV 1000
Query 194 PLSRSCLVVELTITPDFQWD 213
R V +TI D+++D
Sbjct 1001 ESLRLVNSVFITIIEDYKFD 1020
> At4g12010
Length=1219
Score = 29.3 bits (64), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 14/36 (38%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 2 RPRRHLKPTLSDIELLRLFSLSSEFKYMPVREEEKL 37
+ R+++ P L+D E L+LFSL++ P++E E L
Sbjct 336 KGRKYVLPKLNDREALKLFSLNAFSNSFPLKEFEGL 371
Lambda K H
0.324 0.138 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3969005466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40