bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
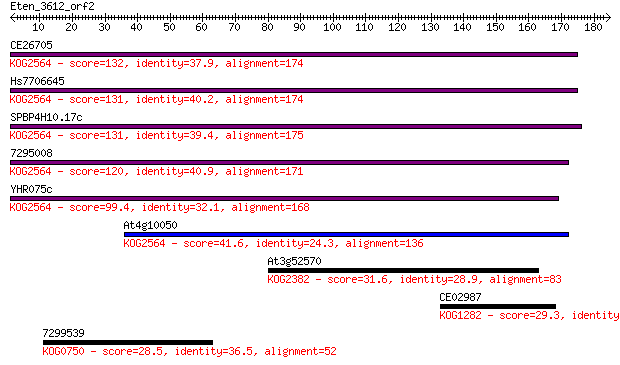
Query= Eten_3612_orf2
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
CE26705 132 3e-31
Hs7706645 131 7e-31
SPBP4H10.17c 131 9e-31
7295008 120 1e-27
YHR075c 99.4 3e-21
At4g10050 41.6 9e-04
At3g52570 31.6 1.00
CE02987 29.3 4.7
7299539 28.5 7.4
> CE26705
Length=364
Score = 132 bits (333), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 66/174 (37%), Positives = 100/174 (57%), Gaps = 17/174 (9%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L+V+DVVEG+A+ AL M F+ PS F S ++A++W + +G N ++A +S+PSQ+
Sbjct 186 LIVIDVVEGSAMEALGGMVHFLHSRPSSFPSIEKAIHWCLSSGTARNPTAARVSMPSQIR 245
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILICS 120
+ + E +TWR+D+ TE YW+GWF G+S FL K+L+ +
Sbjct 246 EVS-----------------EHEYTWRIDLTTTEQYWKGWFEGLSKEFLGCSVPKMLVLA 288
Query 121 SSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYRLHLP 174
DRLD++L I MQGKFQ ++ GH ++ED P L V F R+R+ P
Sbjct 289 GVDRLDRDLTIGQMQGKFQTCVLPKVGHCVQEDSPQNLADEVGRFACRHRIAQP 342
> Hs7706645
Length=386
Score = 131 bits (330), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 70/199 (35%), Positives = 103/199 (51%), Gaps = 25/199 (12%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L ++DVVEGTA+ AL M F+ P F S + A+ WS+ +G + N SA +S+ Q+
Sbjct 177 LCMIDVVEGTAMDALNSMQNFLRGRPKTFKSLENAIEWSVKSGQIRNLESARVSMVGQVK 236
Query 61 KTTRGALPASHKGA-------------------------DKGPPDEEVWTWRVDVMATEP 95
+ P K D + +TWR+++ TE
Sbjct 237 QCEGITSPEGSKSIVEGIIEEEEEDEEGSESISKRKKEDDMETKKDHPYTWRIELAKTEK 296
Query 96 YWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQP 155
YW+GWFRG+S+ FLS K+L+ + DRLDK+L I MQGKFQ+Q++ GH + ED P
Sbjct 297 YWDGWFRGLSNLFLSCPIPKLLLLAGVDRLDKDLTIGQMQGKFQMQVLPQCGHAVHEDAP 356
Query 156 AELCRVVQTFITRYRLHLP 174
++ V TF+ R+R P
Sbjct 357 DKVAEAVATFLIRHRFAEP 375
> SPBP4H10.17c
Length=341
Score = 131 bits (329), Expect = 9e-31, Method: Compositional matrix adjust.
Identities = 69/176 (39%), Positives = 102/176 (57%), Gaps = 18/176 (10%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
L+V+DVVEGTA+ AL M ++S P+ F S +A++W I + NR SA I++PS LV
Sbjct 174 LVVIDVVEGTAMEALGFMKTYLSNRPTSFKSIDDAISWHIKTLVTRNRLSACITVPSLLV 233
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILICS 120
+ G + WR D+ T PYW WF+G+S FL + ++LI +
Sbjct 234 QQEDGT-----------------FVWRTDLYKTSPYWMDWFKGLSDKFLRAPYGRMLIVA 276
Query 121 SSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYR-LHLPP 175
+DRLDK L I MQGK+Q++I+ +GH + ED PA++ ++ F R + L LPP
Sbjct 277 GTDRLDKTLTIGQMQGKYQLEILPETGHFVHEDVPAKISSLLLNFWHRNQPLVLPP 332
> 7295008
Length=402
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 70/224 (31%), Positives = 105/224 (46%), Gaps = 53/224 (23%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
+ V+DVVEGTA+ AL M +F+ P F S A+ W I +G + N SA +S+P Q++
Sbjct 165 ITVIDVVEGTAMEALASMQSFLRSRPKYFQSIPNAIEWCIRSGQVRNVDSAKVSMPGQII 224
Query 61 KTTRGAL-------------PASHK--------------------------------GAD 75
T L A H GAD
Sbjct 225 NCTTNKLATNDLPLPDDVLEEAHHNSMFPNPFSISEDEESSPPGDDAADGSSESAAAGAD 284
Query 76 KGPPDE--------EVWTWRVDVMATEPYWEGWFRGMSHAFLSSRCVKILICSSSDRLDK 127
P+ + +TWR+D+ +E YW GWF G+S FL+ R K L+ +S D LD+
Sbjct 285 FKKPNTTKSTTEAAKNYTWRIDLSKSEKYWVGWFSGLSDKFLNLRLPKQLLLASIDGLDR 344
Query 128 ELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITRYRL 171
L + MQG+FQ+Q+++ GH + ED+P E+ V+ ++ R R
Sbjct 345 TLTVGQMQGRFQMQVLARCGHAVHEDRPHEVAEVISGYLIRNRF 388
> YHR075c
Length=400
Score = 99.4 bits (246), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 54/169 (31%), Positives = 88/169 (52%), Gaps = 19/169 (11%)
Query 1 LMVLDVVEGTALAALPQMAAFVSRFPSLFTSCKEAVNWSIFAGLLCNRSSAAISIPSQLV 60
+ +LD+VE A+ AL ++ F+ P++F S +AV+W + L RSSA I+IP+
Sbjct 229 ITMLDIVEEAAIMALNKVEHFLQNTPNVFESINDAVDWHVQHALSRLRSSAEIAIPALFA 288
Query 61 KTTRGALPASHKGADKGPPDEEVWTWRVDVMAT-EPYWEGWFRGMSHAFLSSRCVKILIC 119
G + R+ + T P+W+ WF +SH+F+ K+LI
Sbjct 289 PLKSGKVV------------------RITNLKTFSPFWDTWFTDLSHSFVGLPVSKLLIL 330
Query 120 SSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQPAELCRVVQTFITR 168
+ ++ LDKEL++ MQGK+Q+ + SGH I+ED P + + F R
Sbjct 331 AGNENLDKELIVGQMQGKYQLVVFQDSGHFIQEDSPIKTAITLIDFWKR 379
> At4g10050
Length=308
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 33/136 (24%), Positives = 56/136 (41%), Gaps = 44/136 (32%)
Query 36 VNWSIFAGLLCNRSSAAISIPSQLVKTTRGALPASHKGADKGPPDEEVWTWRVDVMATEP 95
+ +S+ G L N SA +SIP+ L K + + +R + TE
Sbjct 191 IEYSVRGGSLRNIDSARVSIPTTL----------------KYDDSKHCYVYRTRLEETEQ 234
Query 96 YWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELMIAHMQGKFQVQIVSGSGHVIEEDQP 155
YW+GW+ +C +++I SS + ++ +G I+ED P
Sbjct 235 YWKGWY----------KCDRLMIFMSSLLCNDDI-----RG-------------IQEDVP 266
Query 156 AELCRVVQTFITRYRL 171
E +V FI+R R+
Sbjct 267 EEFANLVLNFISRNRI 282
> At3g52570
Length=343
Score = 31.6 bits (70), Expect = 1.00, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 37/95 (38%), Gaps = 12/95 (12%)
Query 80 DEEVWTWRVD--VMATEPYWEGWFRGMSHAFLSSRCVKILICSSSDRLDKELM-----IA 132
D E WT+ +D V Y E + + + +I SDR D + IA
Sbjct 240 DSETWTFNLDGAVQMFNSYRETSYWSLLENPPKETEINFVIAEKSDRWDNDTTKRLETIA 299
Query 133 HM-----QGKFQVQIVSGSGHVIEEDQPAELCRVV 162
+ +GK ++ SGH + D P L +V
Sbjct 300 NQRQNVAEGKVATHLLRNSGHWVHTDNPKGLLEIV 334
> CE02987
Length=505
Score = 29.3 bits (64), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 12/37 (32%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 133 HMQGKFQVQIVS--GSGHVIEEDQPAELCRVVQTFIT 167
H+ K + +V+ GSGH + D+P ++V F+T
Sbjct 456 HLGSKLSIDVVTVKGSGHFVPLDRPGPALQMVHNFLT 492
> 7299539
Length=240
Score = 28.5 bits (62), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 26/53 (49%), Gaps = 1/53 (1%)
Query 11 ALAALPQMAAFVSRFPSLFTSCKEAV-NWSIFAGLLCNRSSAAISIPSQLVKT 62
++ P MA + P EAV WS+ AGLL +SA + P +VKT
Sbjct 120 SMVYFPLMAWINDQGPRKSDGSGEAVFYWSLIAGLLSGMTSAFMVTPFDVVKT 172
Lambda K H
0.322 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40