bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
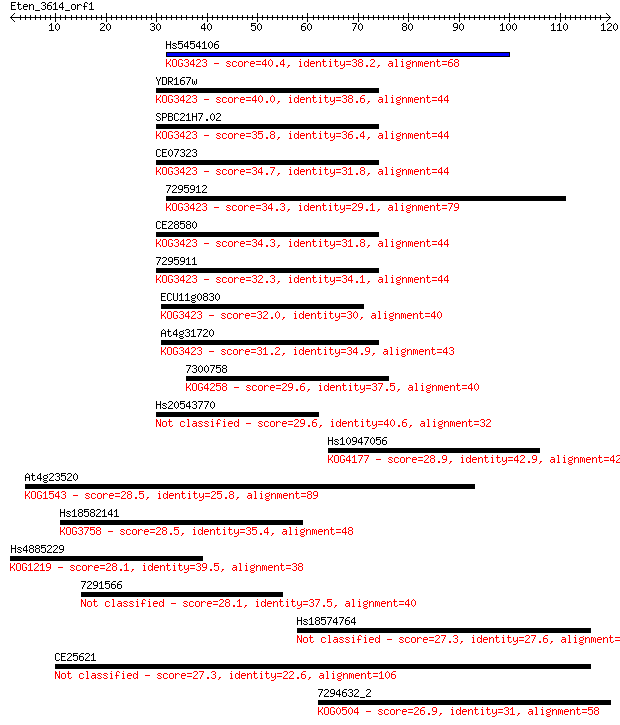
Query= Eten_3614_orf1
Length=119
Score E
Sequences producing significant alignments: (Bits) Value
Hs5454106 40.4 7e-04
YDR167w 40.0 0.001
SPBC21H7.02 35.8 0.022
CE07323 34.7 0.048
7295912 34.3 0.050
CE28580 34.3 0.051
7295911 32.3 0.19
ECU11g0830 32.0 0.30
At4g31720 31.2 0.47
7300758 29.6 1.5
Hs20543770 29.6 1.6
Hs10947056 28.9 2.3
At4g23520 28.5 2.9
Hs18582141 28.5 3.2
Hs4885229 28.1 3.8
7291566 28.1 3.8
Hs18574764 27.3 6.2
CE25621 27.3 7.1
7294632_2 26.9 8.1
> Hs5454106
Length=218
Score = 40.4 bits (93), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 33/68 (48%), Gaps = 1/68 (1%)
Query 32 LLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQVAFEKAIDEAKLILASSR 91
L +L + P I D V YYL+R G +DP RL+S+A Q F I L +
Sbjct 119 FLMQLEDYTPTIPDAVTGYYLNRAGFEASDPRIIRLISLAAQ-KFISDIANDALQHCKMK 177
Query 92 GEASVVSR 99
G AS SR
Sbjct 178 GTASGSSR 185
> YDR167w
Length=206
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 0/44 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
E++L+ + + P I D V+DYYL++ G D RLL++A Q
Sbjct 75 EEILEMMDSTPPIIPDAVIDYYLTKNGFNVADVRVKRLLALATQ 118
> SPBC21H7.02
Length=215
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 0/44 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
E L ++ + P I D ++DYYLS G DP +LL + Q
Sbjct 94 ENFLAQMDDYSPLIPDVLLDYYLSLSGFKCVDPRLKKLLGLTAQ 137
> CE07323
Length=176
Score = 34.7 bits (78), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
+ + +L+ + P I D V ++L G +DP TR++S+A Q
Sbjct 77 HEFINQLADYPPTIPDSVTLHFLKSAGVDGSDPRVTRMISLAAQ 120
> 7295912
Length=146
Score = 34.3 bits (77), Expect = 0.050, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 36/79 (45%), Gaps = 3/79 (3%)
Query 32 LLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQVAFEKAIDEAKLILASSR 91
+ +L + P I D V +YL+ G + D RL+S+A Q ID+A L S+
Sbjct 44 FMSQLEDYTPLIPDAVTSHYLNMGGFQSDDKRIVRLISLAAQKYMSDIIDDA---LQHSK 100
Query 92 GEASVVSRRTDRGSSTQLR 110
+ + T GS + R
Sbjct 101 ARTHMQTTNTPGGSKAKDR 119
> CE28580
Length=179
Score = 34.3 bits (77), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 14/44 (31%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
+ + +L+ + P I D V ++L G +DP TR++S+A Q
Sbjct 80 HEFINQLADYPPTIPDSVTLHFLKSAGVDGSDPRVTRMISLAAQ 123
> 7295911
Length=167
Score = 32.3 bits (72), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 0/44 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
++L+++L + P I D + + L G T DP RL+SV+ Q
Sbjct 58 DELIKQLEDYSPTIPDALTMHILKTAGFCTVDPKIVRLVSVSAQ 101
> ECU11g0830
Length=133
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 12/40 (30%), Positives = 25/40 (62%), Gaps = 0/40 (0%)
Query 31 QLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSV 70
+L Q L ++ P + + V DY++ + G T+D + +L+S+
Sbjct 33 ELKQNLDSYTPLLPESVTDYFMEKAGVVTSDQSVKKLVSL 72
> At4g31720
Length=134
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 31 QLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAATRLLSVAVQ 73
+ L L + P I D++V++YL++ G D RL++VA Q
Sbjct 20 EFLASLMDYTPTIPDDLVEHYLAKSGFQCPDVRLIRLVAVATQ 62
> 7300758
Length=1742
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 15/42 (35%), Positives = 23/42 (54%), Gaps = 2/42 (4%)
Query 36 LSAHEPAILDEVVDYYLSRVGCT--TTDPAATRLLSVAVQVA 75
L AH P DE + YL+R+G T P R+ +A+++A
Sbjct 1463 LRAHRPEERDEAMMTYLNRIGVTGNVQPPTYGRIYQMAIEIA 1504
> Hs20543770
Length=545
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 30 EQLLQKLSAHEPAILDEVVDYYLSRVGCTTTD 61
E L++ A + +DEV+ YYL R GC + D
Sbjct 430 ESLVESCFATPTSKIDEVLKYYLIRDGCVSDD 461
> Hs10947056
Length=4377
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 23/42 (54%), Gaps = 0/42 (0%)
Query 64 ATRLLSVAVQVAFEKAIDEAKLILASSRGEASVVSRRTDRGS 105
AT LL+ A V F D L +AS RG A++V DRG+
Sbjct 251 ATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGA 292
> At4g23520
Length=355
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 23/91 (25%), Positives = 37/91 (40%), Gaps = 2/91 (2%)
Query 4 QNSSDRQSTTMDKSVLDLEEILGPRDEQLLQKLSAHEPAI--LDEVVDYYLSRVGCTTTD 61
Q S +R+ + +D E + DE LQK AH+P +D+ ++ C
Sbjct 226 QGSCNRKQVHLLVITIDSYEDVPANDEISLQKAVAHQPVSVGVDKKSQEFMLYRSCIYNG 285
Query 62 PAATRLLSVAVQVAFEKAIDEAKLILASSRG 92
P T L V V + + I+ +S G
Sbjct 286 PCGTNLDHALVIVGYGSENGQDYWIVRNSWG 316
> Hs18582141
Length=605
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/48 (35%), Positives = 25/48 (52%), Gaps = 0/48 (0%)
Query 11 STTMDKSVLDLEEILGPRDEQLLQKLSAHEPAILDEVVDYYLSRVGCT 58
S M K+ L L E R E L ++ AH +++E Y L+RVG +
Sbjct 452 SLYMMKTTLALFEFTDRRLEMLQFQIEAHLDTLINEQASYVLTRVGLS 499
> Hs4885229
Length=4590
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 1 GTEQNSSDRQSTTMDKSVLDLEEILGPRDEQLLQKLSA 38
G+ Q S+ + ++ SV D+EEI+G R + QKL A
Sbjct 3704 GSAQISTKQLLHKINSSVTDIEEIIGVRILNVFQKLCA 3741
> 7291566
Length=1489
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 28/43 (65%), Gaps = 3/43 (6%)
Query 15 DKSVLDLEEILGPRDEQLLQKLSAHEP-AILDEVV--DYYLSR 54
++ V++L+E + RD QL +K+ A E A DE++ + YL++
Sbjct 1107 NRQVVELQEAMATRDRQLQEKIEASEKLAKFDEILIENEYLNK 1149
> Hs18574764
Length=1275
Score = 27.3 bits (59), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 30/61 (49%), Gaps = 3/61 (4%)
Query 58 TTTDPAATRLLSVAVQVAFEKAIDEAKLILASSRGEASVVSRRTDRGSS---TQLRPMQS 114
T P+ + +V +Q E+++DE + GEA + ++ D +S T+ RP +
Sbjct 444 TVLHPSCLSVCNVTIQDTMERSMDEFTASTPADLGEAGRLRKKADIATSKTTTRFRPSNT 503
Query 115 K 115
K
Sbjct 504 K 504
> CE25621
Length=657
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 24/111 (21%), Positives = 55/111 (49%), Gaps = 5/111 (4%)
Query 10 QSTTMDKSVLDLEEILGPRDEQLLQKLSAHEPAILDEVVDYYLSRVGCTTTDPAA-TRLL 68
+STT + +D E + P + +L H+ IL+E +D + + + D T+ +
Sbjct 46 ESTTPEPLSVDQERDMTPGEHELQVHQKHHQSPILEEEIDVSNTEIDASVLDDHMDTQTM 105
Query 69 SVAVQVAFEKAIDEAKLILASSRGEASVVS----RRTDRGSSTQLRPMQSK 115
+ +++A ++ +A+++L S A+ V+ + R + T+ P Q++
Sbjct 106 EIEMEMALKREKIDAQMVLNSDFSVANEVTISSMKYQGRPTPTRKVPYQNE 156
> 7294632_2
Length=959
Score = 26.9 bits (58), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 24/58 (41%), Gaps = 0/58 (0%)
Query 62 PAATRLLSVAVQVAFEKAIDEAKLILASSRGEASVVSRRTDRGSSTQLRPMQSKCSLV 119
P + LL V + LILA+ RG VV GSS + +C+LV
Sbjct 451 PMVSLLLEFGADVGLTNSQGCTPLILAAMRGHCDVVRPLVAAGSSLGQLDITQRCALV 508
Lambda K H
0.311 0.126 0.330
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40