bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3634_orf1
Length=132
Score E
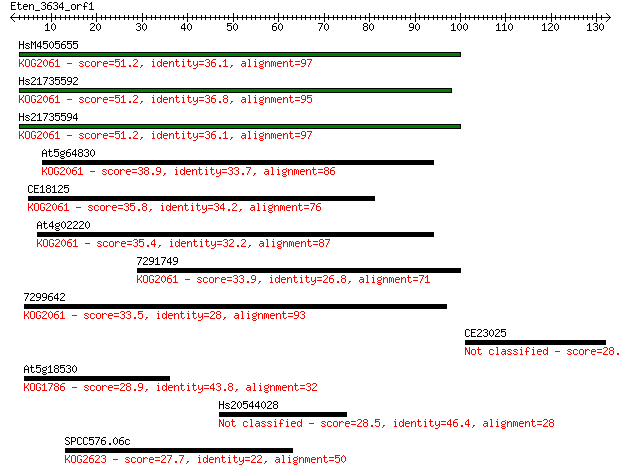
Sequences producing significant alignments: (Bits) Value
HsM4505655 51.2 5e-07
Hs21735592 51.2 5e-07
Hs21735594 51.2 6e-07
At5g64830 38.9 0.003
CE18125 35.8 0.022
At4g02220 35.4 0.029
7291749 33.9 0.088
7299642 33.5 0.11
CE23025 28.9 2.5
At5g18530 28.9 2.8
Hs20544028 28.5 3.5
SPCC576.06c 27.7 5.7
> HsM4505655
Length=344
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 45/102 (44%), Gaps = 10/102 (9%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVC 61
A PV +GF ES P + + SK GG P W+ +P C LCG L F+
Sbjct 4 AGARPVELGF-AESAPAWRLRSEQFPSKVGGRPAWLGAAGLPGPQALACELCGRPLSFLL 62
Query 62 QVATPY----GANKRCLYFFCCHAQATCGKRPEGWKVLRGVV 99
QV P A RC++ FCC Q C G +V R +
Sbjct 63 QVYAPLPGRPDAFHRCIFLFCCREQPCCA----GLRVFRNQL 100
> Hs21735592
Length=344
Score = 51.2 bits (121), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 35/100 (35%), Positives = 44/100 (44%), Gaps = 10/100 (10%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVC 61
A PV +GF ES P + + SK GG P W+ +P C LCG L F+
Sbjct 4 AGARPVELGF-AESAPAWRLRSEQFPSKVGGRPAWLGAAGLPGPQALACELCGRPLSFLL 62
Query 62 QVATPY----GANKRCLYFFCCHAQATCGKRPEGWKVLRG 97
QV P A RC++ FCC Q C G +V R
Sbjct 63 QVYAPLPGRPDAFHRCIFLFCCREQPCCA----GLRVFRN 98
> Hs21735594
Length=228
Score = 51.2 bits (121), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 35/102 (34%), Positives = 45/102 (44%), Gaps = 10/102 (9%)
Query 3 ASETPVLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVC 61
A PV +GF ES P + + SK GG P W+ +P C LCG L F+
Sbjct 4 AGARPVELGF-AESAPAWRLRSEQFPSKVGGRPAWLGAAGLPGPQALACELCGRPLSFLL 62
Query 62 QVATPY----GANKRCLYFFCCHAQATCGKRPEGWKVLRGVV 99
QV P A RC++ FCC Q C G +V R +
Sbjct 63 QVYAPLPGRPDAFHRCIFLFCCREQPCCA----GLRVFRNQL 100
> At5g64830
Length=380
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 38/95 (40%), Gaps = 12/95 (12%)
Query 8 VLIGFFGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFP----FNCSLCGVVLDFVCQV 63
+L+G G+ D + + +K GG P W IPD NC CG L V QV
Sbjct 4 ILLGLPGKWAEDELEPSDHYTTKIGGLPDW--PPIPDDALKPELLNCCSCGSKLSLVAQV 61
Query 64 ATPYGA-----NKRCLYFFCCHAQATCGKRPEGWK 93
P +R LY F C CG + W+
Sbjct 62 YAPISTEILDIQERTLYIFGC-LMPKCGTSEQSWR 95
> CE18125
Length=386
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 38/86 (44%), Gaps = 10/86 (11%)
Query 5 ETPVLIGFFGESVPDGDQTAAEE---LSKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFV 60
+ PV +GF + PD + L K GG P W++ K +P C++C L F+
Sbjct 7 DEPVNLGFGVQFEPDDLYRLRSQFLPLGKIGGKPSWLNPKNLPKSADLLCNVCEKPLCFL 66
Query 61 CQVATPYGAN------KRCLYFFCCH 80
QV+ G N R L+ F C
Sbjct 67 MQVSANGGINDPPHAFHRSLFLFVCR 92
> At4g02220
Length=446
Score = 35.4 bits (80), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 16/98 (16%)
Query 7 PVLIGFFGESVPDGDQTAAEELSKF-----GGNPVWIDG-KIPDGFPFNCSLCGVVLDFV 60
PV++GF + + A L + GG P W+D +P G C LC + FV
Sbjct 79 PVMLGFV-----ESPKFAWSNLRQLFPNLAGGVPAWLDPVNLPSGKSILCDLCEEPMQFV 133
Query 61 CQVATPY----GANKRCLYFFCCHAQATCGK-RPEGWK 93
Q+ P A R L+ F C + + + + E WK
Sbjct 134 LQLYAPLTDKESAFHRTLFLFMCPSMSCLLRDQHEQWK 171
> 7291749
Length=347
Score = 33.9 bits (76), Expect = 0.088, Method: Compositional matrix adjust.
Identities = 19/76 (25%), Positives = 32/76 (42%), Gaps = 5/76 (6%)
Query 29 SKFGGNPVWIDGK-IPDGFPFNCSLCGVVLDFVCQVATP----YGANKRCLYFFCCHAQA 83
SK GG P W++ + +P CS C F+ Q+ P Y ++ F C ++
Sbjct 24 SKLGGQPAWLELEALPPTSQLQCSKCRAPKSFLAQLYAPFEDEYNFHRSIYVFLCRNSDC 83
Query 84 TCGKRPEGWKVLRGVV 99
+ + VLR +
Sbjct 84 QEAQNASNFTVLRSQL 99
> 7299642
Length=485
Score = 33.5 bits (75), Expect = 0.11, Method: Composition-based stats.
Identities = 26/96 (27%), Positives = 43/96 (44%), Gaps = 7/96 (7%)
Query 4 SETPVLIGFFGESV-PDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQ 62
+++ V +G+ E V +Q +K GG P W ++ P +C LCG V + Q
Sbjct 4 NKSTVYLGYEDEEVTAKQEQLLNSCTNKIGGTPDWPRHEVT--IP-SCPLCGAVRPLIVQ 60
Query 63 VATPYGANK--RCLYFFCCHAQATCGKRPEGWKVLR 96
+ P ++ R LY F C C + + W +R
Sbjct 61 MYAPLDRSQFHRSLYVFGC-MNPVCSQNSKSWCCVR 95
> CE23025
Length=540
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 17/31 (54%), Gaps = 0/31 (0%)
Query 101 PSAPAQAEAEAYGAAHTSFKRENDDLAAADP 131
P+APAQ E+ A F E+D+ A DP
Sbjct 504 PAAPAQESTESSAAQKREFLPEHDEDEAQDP 534
> At5g18530
Length=1598
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 4 SETPVLIGFFGESVPDGDQTAAEELSKFGGNP 35
S+T LI F ES D DQ +++ SK NP
Sbjct 1324 SQTGKLISLFSESPSDQDQASSDPSSKNNSNP 1355
> Hs20544028
Length=1200
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 47 PFNCSLCGVVLDFVCQVATPYGANKRCL 74
PFN +L G+ FV TP G +RCL
Sbjct 493 PFNQTLTGLHAAFVFHQLTPRGVVRRCL 520
> SPCC576.06c
Length=445
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 11/50 (22%), Positives = 22/50 (44%), Gaps = 0/50 (0%)
Query 13 FGESVPDGDQTAAEELSKFGGNPVWIDGKIPDGFPFNCSLCGVVLDFVCQ 62
+ + P ++ ++L K GN +W+D K+ D + D C+
Sbjct 231 YALTTPLLTSSSGQKLGKSAGNAIWLDPKLTDSYSLYQYFISAPDDLACK 280
Lambda K H
0.320 0.137 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40