bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3651_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
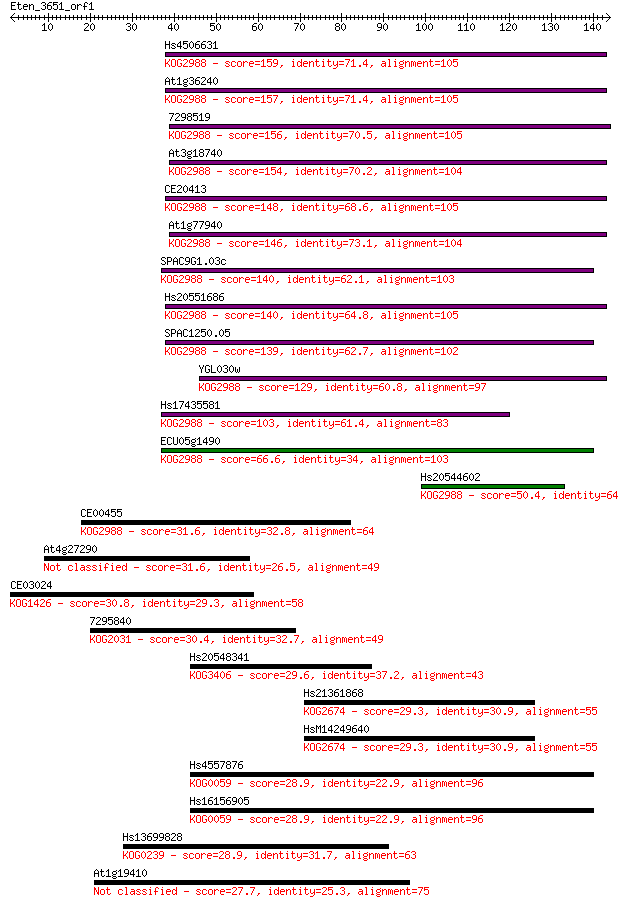
Hs4506631 159 2e-39
At1g36240 157 7e-39
7298519 156 1e-38
At3g18740 154 6e-38
CE20413 148 3e-36
At1g77940 146 1e-35
SPAC9G1.03c 140 7e-34
Hs20551686 140 1e-33
SPAC1250.05 139 3e-33
YGL030w 129 2e-30
Hs17435581 103 8e-23
ECU05g1490 66.6 2e-11
Hs20544602 50.4 1e-06
CE00455 31.6 0.47
At4g27290 31.6 0.56
CE03024 30.8 0.92
7295840 30.4 1.1
Hs20548341 29.6 2.0
Hs21361868 29.3 2.4
HsM14249640 29.3 2.5
Hs4557876 28.9 3.3
Hs16156905 28.9 3.3
Hs13699828 28.9 3.4
At1g19410 27.7 6.9
> Hs4506631
Length=115
Score = 159 bits (402), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 86/105 (81%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ E INSRLQLVMKSGK LG K + +R GKAKL+I++NNCPALRKSEIEYY
Sbjct 4 AKKTKKSLESINSRLQLVMKSGKYVLGYKQTLKMIRQGKAKLVILANNCPALRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++ DPGDSDIIRS+
Sbjct 64 AMLAKTGVHHYSGNNIELGTACGKYYRVCTLAIIDPGDSDIIRSM 108
> At1g36240
Length=112
Score = 157 bits (397), Expect = 7e-39, Method: Compositional matrix adjust.
Identities = 75/105 (71%), Positives = 87/105 (82%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
AKK K+ EGINSRL LVMKSGK LG K+ + SLR+ K KLI+IS+NCP LR+SEIEYY
Sbjct 4 AKKTKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRSSKGKLILISSNCPPLRRSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY +N +LGTACGK FRVSCLS+ DPGDSDII+S+
Sbjct 64 AMLAKVGVHHYNRNNVDLGTACGKYFRVSCLSIVDPGDSDIIKSL 108
> 7298519
Length=111
Score = 156 bits (395), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 74/105 (70%), Positives = 84/105 (80%), Gaps = 0/105 (0%)
Query 39 KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYA 98
KKQK+ E N+RL LVMKSGK CLG K + +LR GKAKL++I++N PALRKSEIEYYA
Sbjct 5 KKQKKALESTNARLALVMKSGKYCLGYKQTLKTLRQGKAKLVLIASNTPALRKSEIEYYA 64
Query 99 MLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSVE 143
MLAK V HY G N ELGTACGK FRV LS+TDPGDSDIIRS+E
Sbjct 65 MLAKTEVQHYSGTNIELGTACGKYFRVCTLSITDPGDSDIIRSLE 109
> At3g18740
Length=112
Score = 154 bits (389), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 73/104 (70%), Positives = 86/104 (82%), Gaps = 0/104 (0%)
Query 39 KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYA 98
KK K+ EGINSRL LVMKSGK LG K+ + SLR+ K KLI+IS+NCP LR+SEIEYYA
Sbjct 5 KKAKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRSSKGKLILISSNCPPLRRSEIEYYA 64
Query 99 MLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
MLAK GVH Y G+N +LGTACGK FRVSCLS+ DPGDSDII+++
Sbjct 65 MLAKVGVHRYNGNNVDLGTACGKYFRVSCLSIVDPGDSDIIKTL 108
> CE20413
Length=113
Score = 148 bits (374), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 72/105 (68%), Positives = 83/105 (79%), Gaps = 0/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
A K ++ AE INSRL +VMK+G+ LG K + SL GKAKL+II+NN P LRKSEIEYY
Sbjct 4 AAKPQKNAENINSRLSMVMKTGQYVLGYKQTLKSLLNGKAKLVIIANNTPPLRKSEIEYY 63
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACG+LFRV L+VTD GDSDII SV
Sbjct 64 AMLAKTGVHHYNGNNIELGTACGRLFRVCTLAVTDAGDSDIILSV 108
> At1g77940
Length=308
Score = 146 bits (369), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 76/126 (60%), Positives = 85/126 (67%), Gaps = 22/126 (17%)
Query 39 KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYA 98
KK K+ EGINSRL LVMKSGK LG K+ + SLR K KLI+IS NCP LR+SEIEYYA
Sbjct 5 KKTKKSHEGINSRLALVMKSGKYTLGYKSVLKSLRGSKGKLILISTNCPPLRRSEIEYYA 64
Query 99 MLAKCGVHHYPG----------------------DNNELGTACGKLFRVSCLSVTDPGDS 136
MLAK GVHHY G DN +LGTACGK FRVSCLS+ DPGDS
Sbjct 65 MLAKVGVHHYNGSKFLSSLARPVFKDSLFSCFCEDNVDLGTACGKYFRVSCLSIVDPGDS 124
Query 137 DIIRSV 142
DII+S+
Sbjct 125 DIIKSI 130
> SPAC9G1.03c
Length=109
Score = 140 bits (354), Expect = 7e-34, Method: Compositional matrix adjust.
Identities = 64/103 (62%), Positives = 81/103 (78%), Gaps = 0/103 (0%)
Query 37 MAKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEY 96
+ KK K+ + IN++L L MKSGK LG K+ + +LR+GKAKLI+I+ NCP LRKSE+EY
Sbjct 5 VTKKSKKSGDTINAKLALTMKSGKYVLGYKSTLKTLRSGKAKLILIAGNCPPLRKSELEY 64
Query 97 YAMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
YAML+K VHHY G N +LGTACGKLFRV L++TD GDSDI+
Sbjct 65 YAMLSKANVHHYAGTNIDLGTACGKLFRVGVLAITDAGDSDIL 107
> Hs20551686
Length=243
Score = 140 bits (352), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 68/105 (64%), Positives = 80/105 (76%), Gaps = 1/105 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
K+ K E IN RLQL MKSGK LG K + +R G AKL+I++ NCPALRKSE+EYY
Sbjct 133 TKEMKNSLESINYRLQL-MKSGKYMLGYKQTLKMIRQGTAKLVILAYNCPALRKSEVEYY 191
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
AMLAK GVHHY G+N ELGTACGK +RV L++TDPGDS IIRS+
Sbjct 192 AMLAKIGVHHYSGNNIELGTACGKYYRVCTLALTDPGDSAIIRSM 236
> SPAC1250.05
Length=117
Score = 139 bits (349), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 64/102 (62%), Positives = 81/102 (79%), Gaps = 0/102 (0%)
Query 38 AKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYY 97
+KK K+ + INS+L L MKSGK LG K+ + +LR+GKAKLI+I+ N P LRKSE+EYY
Sbjct 14 SKKGKKSGDTINSKLALTMKSGKYVLGYKSTLKTLRSGKAKLILIAANAPPLRKSELEYY 73
Query 98 AMLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
AML++C VHHY G+N +LGTACGKLFRV L+V D GDSDI+
Sbjct 74 AMLSRCSVHHYSGNNIDLGTACGKLFRVGVLAVIDAGDSDIL 115
> YGL030w
Length=105
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 59/97 (60%), Positives = 75/97 (77%), Gaps = 0/97 (0%)
Query 46 EGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKCGV 105
E IN +L LV+KSGK LG K+ V SLR GK+KLIII+ N P LRKSE+EYYAML+K V
Sbjct 8 ESINQKLALVIKSGKYTLGYKSTVKSLRQGKSKLIIIAANTPVLRKSELEYYAMLSKTKV 67
Query 106 HHYPGDNNELGTACGKLFRVSCLSVTDPGDSDIIRSV 142
+++ G NNELGTA GKLFRV +S+ + GDSDI+ ++
Sbjct 68 YYFQGGNNELGTAVGKLFRVGVVSILEAGDSDILTTL 104
> Hs17435581
Length=169
Score = 103 bits (258), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 51/83 (61%), Positives = 58/83 (69%), Gaps = 0/83 (0%)
Query 37 MAKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEY 96
+AKK K+ E INSRL LV+KSGK L K +R GKAKL I + NCPALRKS+IEY
Sbjct 3 LAKKIKKSVEWINSRLLLVIKSGKYILAYKQTPKMIRQGKAKLAIHTENCPALRKSKIEY 62
Query 97 YAMLAKCGVHHYPGDNNELGTAC 119
Y ML K GVHHY +N ELG AC
Sbjct 63 YTMLTKTGVHHYSDNNFELGIAC 85
> ECU05g1490
Length=108
Score = 66.6 bits (161), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 35/104 (33%), Positives = 64/104 (61%), Gaps = 2/104 (1%)
Query 37 MAKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEY 96
M K +G + + S L L M++GK +G K A+ S+ KAK ++IS+N P+ ++ +EY
Sbjct 1 MRGKSNKG-DNVTSSLLLAMRTGKYIVGFKRAIKSVIMKKAKCLVISSNFPSTKRKLLEY 59
Query 97 YAMLA-KCGVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
Y++LA + + +N ELG+ + R+ C+S+ D G++++I
Sbjct 60 YSVLAGGLPIIFHSANNEELGSITERGHRLGCISIIDQGEAELI 103
> Hs20544602
Length=165
Score = 50.4 bits (119), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/34 (64%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 99 MLAKCGVHHYPGDNNELGTACGKLFRVSCLSVTD 132
MLAK GVHHY G+N ELGTACGK +RV ++ D
Sbjct 1 MLAKTGVHHYGGNNIELGTACGKYYRVGTPAIID 34
> CE00455
Length=178
Score = 31.6 bits (70), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 6/69 (8%)
Query 18 FRPSAAAAAAAARAVCTPKMA-----KKQKQGAEGINSRLQLVMKSGKVCLGLKTAVHSL 72
F P + RA+ + + AE IN RL +VMK+G+ L + + SL
Sbjct 68 FFPFTSILFLIYRAIIYNVYSGHTGIFSKFTNAENINFRLSMVMKTGQYVL-YEQKLKSL 126
Query 73 RTGKAKLII 81
AKL+I
Sbjct 127 LNENAKLVI 135
> At4g27290
Length=772
Score = 31.6 bits (70), Expect = 0.56, Method: Composition-based stats.
Identities = 13/49 (26%), Positives = 24/49 (48%), Gaps = 0/49 (0%)
Query 9 APGKFGKGKFRPSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMK 57
A K G+G F P A + V ++++ +QG E + ++L+ K
Sbjct 456 AGNKLGQGGFGPVYKGTLACGQEVAVKRLSRTSRQGVEEFKNEIKLIAK 504
> CE03024
Length=569
Score = 30.8 bits (68), Expect = 0.92, Method: Composition-based stats.
Identities = 17/58 (29%), Positives = 30/58 (51%), Gaps = 2/58 (3%)
Query 1 STSNGVSIAPGKFGKGKFRPSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMKS 58
S + V P + G+ K PS + AAA + TP ++++G+E +N+ + M S
Sbjct 65 SIVDNVPKTPRRAGRPKKLPSTSTAAA--ESAVTPGKRGRKRKGSESVNTTINSTMNS 120
> 7295840
Length=580
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 16/49 (32%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 20 PSAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMKSGKVCLGLKTA 68
PS ++A+ A + T +AKKQK A+ I + +V++ G + L+ A
Sbjct 99 PSGSSASGPAASQDTSNLAKKQKLNAKNIRDYIPVVIEKGGMAKKLERA 147
> Hs20548341
Length=110
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query 44 GAEGINSRLQLVMKSGKVCLGLKTAVHS----LRTGKAKLIIISNNC 86
G IN+ LQ V+K+ + GL +H L G+A L ++++NC
Sbjct 10 GVMDINTALQEVLKTTLIHDGLARGIHEAAKPLDKGQAHLYVLASNC 56
> Hs21361868
Length=330
Score = 29.3 bits (64), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 71 SLRTGKAKLIIISNNCPALRKSEIEYYAMLAKCGVHHYPGDNNELGTACGKLFRV 125
++ +A + S +C + RK + + C V Y GD E GT C +L RV
Sbjct 219 TVDVSQADFPLESFHCTSPRK--MAFAKTDPSCTVGFYAGDRKEFGTLCSELTRV 271
> HsM14249640
Length=141
Score = 29.3 bits (64), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 26/55 (47%), Gaps = 2/55 (3%)
Query 71 SLRTGKAKLIIISNNCPALRKSEIEYYAMLAKCGVHHYPGDNNELGTACGKLFRV 125
++ +A + S +C + RK + + M C V Y GD E T C +L RV
Sbjct 30 TVDVSQADFPLESFHCTSPRK--MAFAKMDPSCTVGFYAGDRKEFETLCSELTRV 82
> Hs4557876
Length=2273
Score = 28.9 bits (63), Expect = 3.3, Method: Composition-based stats.
Identities = 22/96 (22%), Positives = 39/96 (40%), Gaps = 2/96 (2%)
Query 44 GAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKC 103
G I + L V +S +C H L + L + ++++E AML
Sbjct 990 GGRDIETSLDAVRQSLGMCPQHNILFHHLTVAEHMLFYAQLKGKSQEEAQLEMEAMLEDT 1049
Query 104 GVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
G+HH N E G + R +++ GD+ ++
Sbjct 1050 GLHH--KRNEEAQDLSGGMQRKLSVAIAFVGDAKVV 1083
> Hs16156905
Length=2273
Score = 28.9 bits (63), Expect = 3.3, Method: Composition-based stats.
Identities = 22/96 (22%), Positives = 39/96 (40%), Gaps = 2/96 (2%)
Query 44 GAEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNCPALRKSEIEYYAMLAKC 103
G I + L V +S +C H L + L + ++++E AML
Sbjct 990 GGRDIETSLDAVRQSLGMCPQHNILFHHLTVAEHMLFYAQLKGKSQEEAQLEMEAMLEDT 1049
Query 104 GVHHYPGDNNELGTACGKLFRVSCLSVTDPGDSDII 139
G+HH N E G + R +++ GD+ ++
Sbjct 1050 GLHH--KRNEEAQDLSGGMQRKLSVAIAFVGDAKVV 1083
> Hs13699828
Length=332
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 11/64 (17%)
Query 28 AARAVCTPKMAKKQKQG-AEGINSRLQLVMKSGKVCLGLKTAVHSLRTGKAKLIIISNNC 86
A++ P++ G AE + +RLQLV +G C+G G AKL++I
Sbjct 240 ASQGALAPQLVPGNPAGHAEQVQARLQLVDSAGSECVG----------GDAKLLVILCIS 289
Query 87 PALR 90
P+ R
Sbjct 290 PSQR 293
> At1g19410
Length=435
Score = 27.7 bits (60), Expect = 6.9, Method: Composition-based stats.
Identities = 19/78 (24%), Positives = 32/78 (41%), Gaps = 3/78 (3%)
Query 21 SAAAAAAAARAVCTPKMAKKQKQGAEGINSRLQLVMKSGKVCLGLKTAVH---SLRTGKA 77
S A R C P + QG ++ QL+ +S VC ++H + + +
Sbjct 110 SYAILMVVPRMACLPSLKNLLLQGVRYVDESFQLLDESHTVCFPSLKSLHLKEVISSDER 169
Query 78 KLIIISNNCPALRKSEIE 95
L + NCP L + +E
Sbjct 170 YLRSLLANCPVLEELVME 187
Lambda K H
0.317 0.131 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40