bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3660_orf1
Length=108
Score E
Sequences producing significant alignments: (Bits) Value
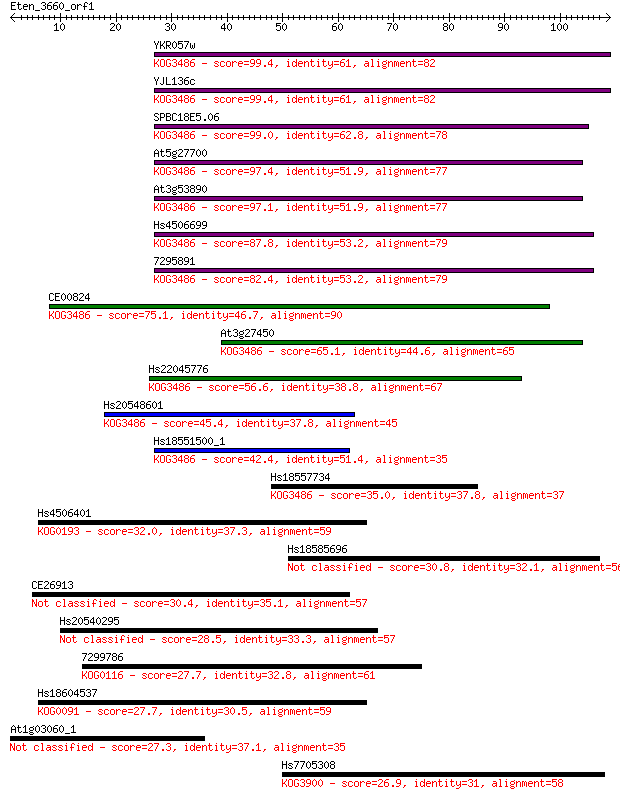
YKR057w 99.4 1e-21
YJL136c 99.4 2e-21
SPBC18E5.06 99.0 2e-21
At5g27700 97.4 5e-21
At3g53890 97.1 7e-21
Hs4506699 87.8 4e-18
7295891 82.4 2e-16
CE00824 75.1 3e-14
At3g27450 65.1 3e-11
Hs22045776 56.6 1e-08
Hs20548601 45.4 3e-05
Hs18551500_1 42.4 2e-04
Hs18557734 35.0 0.037
Hs4506401 32.0 0.29
Hs18585696 30.8 0.63
CE26913 30.4 0.92
Hs20540295 28.5 3.0
7299786 27.7 5.2
Hs18604537 27.7 5.3
At1g03060_1 27.3 6.5
Hs7705308 26.9 9.2
> YKR057w
Length=87
Score = 99.4 bits (246), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 50/87 (57%), Positives = 63/87 (72%), Gaps = 5/87 (5%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYT-GEYTTVAISGAVR 85
MEND+G++V+LYVPRKCSAT+R+I A +H +VQINV VDE GR GEY T A+SG VR
Sbjct 1 MENDKGQLVELYVPRKCSATNRIIKADDHASVQINVAKVDEEGRAIPGEYVTYALSGYVR 60
Query 86 QRGESDACMNRLM----FEKKILSFSK 108
RGESD +NRL K + S+S+
Sbjct 61 SRGESDDSLNRLAQNDGLLKNVWSYSR 87
> YJL136c
Length=87
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 50/87 (57%), Positives = 63/87 (72%), Gaps = 5/87 (5%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYT-GEYTTVAISGAVR 85
MEND+G++V+LYVPRKCSAT+R+I A +H +VQINV VDE GR GEY T A+SG VR
Sbjct 1 MENDKGQLVELYVPRKCSATNRIIKADDHASVQINVAKVDEEGRAIPGEYITYALSGYVR 60
Query 86 QRGESDACMNRLM----FEKKILSFSK 108
RGESD +NRL K + S+S+
Sbjct 61 SRGESDDSLNRLAQNDGLLKNVWSYSR 87
> SPBC18E5.06
Length=87
Score = 99.0 bits (245), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 49/79 (62%), Positives = 60/79 (75%), Gaps = 1/79 (1%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYT-GEYTTVAISGAVR 85
MEN+ G++VDLYVPRKCSAT+R+I AK+H +VQINV VD GR GE TT AISG VR
Sbjct 1 MENEAGQLVDLYVPRKCSATNRIIQAKDHASVQINVCAVDAEGRQIPGEKTTYAISGFVR 60
Query 86 QRGESDACMNRLMFEKKIL 104
+GESD C+NRL + +L
Sbjct 61 SKGESDDCINRLTTQDGLL 79
> At5g27700
Length=85
Score = 97.4 bits (241), Expect = 5e-21, Method: Compositional matrix adjust.
Identities = 40/77 (51%), Positives = 63/77 (81%), Gaps = 0/77 (0%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYTGEYTTVAISGAVRQ 86
M+N+EG+V +LY+PRKCSAT+RLI +K+H +VQ+N+G +D G YTG++TT A+ G VR
Sbjct 1 MQNEEGQVTELYIPRKCSATNRLITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRA 60
Query 87 RGESDACMNRLMFEKKI 103
+G++D+ ++RL +KK+
Sbjct 61 QGDADSGVDRLWQKKKV 77
> At3g53890
Length=82
Score = 97.1 bits (240), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 40/77 (51%), Positives = 62/77 (80%), Gaps = 0/77 (0%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYTGEYTTVAISGAVRQ 86
MEND G+V +LY+PRKCSAT+R+I +K+H +VQ+N+G +D G YTG++TT A+ G VR
Sbjct 1 MENDAGQVTELYIPRKCSATNRMITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRA 60
Query 87 RGESDACMNRLMFEKKI 103
+G++D+ ++RL +KK+
Sbjct 61 QGDADSGVDRLWQKKKV 77
> Hs4506699
Length=83
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 42/80 (52%), Positives = 58/80 (72%), Gaps = 1/80 (1%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDE-AGRYTGEYTTVAISGAVR 85
M+ND G VDLYVPRKCSA++R+I AK+H ++Q+NV VD+ GR+ G++ T AI GA+R
Sbjct 1 MQNDAGEFVDLYVPRKCSASNRIIGAKDHASIQMNVAEVDKVTGRFNGQFKTYAICGAIR 60
Query 86 QRGESDACMNRLMFEKKILS 105
+ GESD + RL I+S
Sbjct 61 RMGESDDSILRLAKADGIVS 80
> 7295891
Length=83
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 42/80 (52%), Positives = 56/80 (70%), Gaps = 1/80 (1%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVD-EAGRYTGEYTTVAISGAVR 85
MEND G VDLYVPRKCSA++R+I AK+H +VQ+++ VD E GR T T AI G +R
Sbjct 1 MENDAGENVDLYVPRKCSASNRIIHAKDHASVQLSIVDVDPETGRQTDGSKTYAICGEIR 60
Query 86 QRGESDACMNRLMFEKKILS 105
+ GESD C+ RL + I++
Sbjct 61 RMGESDDCIVRLAKKDGIIT 80
> CE00824
Length=151
Score = 75.1 bits (183), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 42/92 (45%), Positives = 59/92 (64%), Gaps = 2/92 (2%)
Query 8 DLSSFISFLLLQDFSKPRKMENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVD- 66
D S + + +Q F M+ND G+ V+LYVPRKCS+++R+I K+H +VQI+ VD
Sbjct 45 DFSDLLVTINVQKFWFQVIMQNDAGQTVELYVPRKCSSSNRIIGPKDHASVQIDFVDVDP 104
Query 67 EAGRYT-GEYTTVAISGAVRQRGESDACMNRL 97
E GR G+ T AI GA+R+ GESD + RL
Sbjct 105 ETGRMIPGKSTRYAICGAIRRMGESDDAILRL 136
> At3g27450
Length=79
Score = 65.1 bits (157), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 29/69 (42%), Positives = 49/69 (71%), Gaps = 4/69 (5%)
Query 39 VPRKC----SATSRLIPAKEHGAVQINVGMVDEAGRYTGEYTTVAISGAVRQRGESDACM 94
+PR+ S T+R I +K+H +VQ+N+G +D G YTG++TT A+ G VR +G++D+ +
Sbjct 6 IPRQSMHIRSTTNRFITSKDHASVQLNIGHLDANGLYTGQFTTFALCGFVRAQGDADSGV 65
Query 95 NRLMFEKKI 103
+RL +KK+
Sbjct 66 DRLRQKKKV 74
> Hs22045776
Length=217
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 45/68 (66%), Gaps = 1/68 (1%)
Query 26 KMENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDE-AGRYTGEYTTVAISGAV 84
+M+ND G + LY+P K S ++ +I AK+H ++Q+N VD+ AGR+ ++ T AI A+
Sbjct 17 EMQNDTGEFIGLYMPWKWSNSNCIIDAKDHKSIQMNGAKVDKVAGRFNHQFKTYAICRAI 76
Query 85 RQRGESDA 92
R+ G + +
Sbjct 77 RRMGRTGS 84
> Hs20548601
Length=147
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 32/45 (71%), Gaps = 0/45 (0%)
Query 18 LQDFSKPRKMENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINV 62
L++ P +N+ ++VDLY+P KCS++ +I AK+H ++Q+N+
Sbjct 36 LRNGRPPGSAKNNASKIVDLYIPWKCSSSHHVIGAKDHTSMQMNM 80
> Hs18551500_1
Length=37
Score = 42.4 bits (98), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 27 MENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQIN 61
M+ND G VDLYVP K S ++ +I AK+H ++Q N
Sbjct 1 MQNDAGEFVDLYVPWKWSDSNCIIDAKDHESIQKN 35
> Hs18557734
Length=160
Score = 35.0 bits (79), Expect = 0.037, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 24/38 (63%), Gaps = 1/38 (2%)
Query 48 RLIPAKEHGAVQINVGMVDE-AGRYTGEYTTVAISGAV 84
R + +H ++Q+NV D+ GR+ G++ T AI GA+
Sbjct 99 RRVRGPDHASIQMNVAEADKVTGRFNGQFKTYAICGAI 136
> Hs4506401
Length=648
Score = 32.0 bits (71), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 5/60 (8%)
Query 6 LFDLSSFISFLLLQDFSKPRKMENDEGRVVDLYVPRKCSATSRL-IPAKEHGAVQINVGM 64
+FD SS IS ++Q F R+ +D+G++ D P K S T R+ +P K+ V + GM
Sbjct 21 VFDGSSCISPTIVQQFGYQRR-ASDDGKLTD---PSKTSNTIRVFLPNKQRTVVNVRNGM 76
> Hs18585696
Length=201
Score = 30.8 bits (68), Expect = 0.63, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 27/56 (48%), Gaps = 8/56 (14%)
Query 51 PAKEHGAVQINVGMVDEAGRYTGEYTTVAISGAVRQRGESDACMNRLMFEKKILSF 106
PA H + E G T Y V ++G+ R+RGE + N+ + KK+ SF
Sbjct 114 PAYAHAII--------ERGFKTMAYVPVPLTGSERRRGEKSSNENKSWYWKKVESF 161
> CE26913
Length=267
Score = 30.4 bits (67), Expect = 0.92, Method: Compositional matrix adjust.
Identities = 20/60 (33%), Positives = 31/60 (51%), Gaps = 5/60 (8%)
Query 5 NLFDLSSFISFLLLQDFSKPRKMEND---EGRVVDLYVPRKCSATSRLIPAKEHGAVQIN 61
N+F + SF LL S RKME+D + R VD + + S R + EHG++ ++
Sbjct 158 NIFLMKDEPSFCLLTSRSHARKMESDKIMDSRAVD--ISQLGSVLFRCLTGHEHGSINLD 215
> Hs20540295
Length=366
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 19/63 (30%), Positives = 30/63 (47%), Gaps = 10/63 (15%)
Query 10 SSFISFLLLQDFSKPRKMENDEGRVVDLYVP------RKCSATSRLIPAKEHGAVQINVG 63
+S SF DF +E+D+G +D +P AT R+I H V++N G
Sbjct 235 TSITSF----DFGGISILEDDDGNSIDDKLPVLSLDAAMAFATGRIILEHVHHVVEVNEG 290
Query 64 MVD 66
++D
Sbjct 291 VID 293
> 7299786
Length=690
Score = 27.7 bits (60), Expect = 5.2, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 30/61 (49%), Gaps = 1/61 (1%)
Query 14 SFLLLQDFSKPRKMENDEGRVVDLYVPRKCSATSRLIPAKEHGAVQINVGMVDEAGRYTG 73
+F+L K + ND R DLY+ + SR +EH VQ+ G VD+A + G
Sbjct 109 TFVLAAQSPKKYYVHNDIFRYQDLYIEDEQDGESRSENDEEHD-VQVVGGTVDQAVQVAG 167
Query 74 E 74
+
Sbjct 168 D 168
> Hs18604537
Length=217
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 1/60 (1%)
Query 6 LFDLSSFISFLLLQDFSKPRKMENDEGRVVDLYVPRKCS-ATSRLIPAKEHGAVQINVGM 64
+FD+++ SF ++D+ + KM R+V L V KC A+ R + +E + + GM
Sbjct 92 VFDITNRRSFEHVKDWLEEAKMYVQPFRIVFLLVGHKCDLASQRQVTREEAEKLSADCGM 151
> At1g03060_1
Length=2884
Score = 27.3 bits (59), Expect = 6.5, Method: Compositional matrix adjust.
Identities = 13/39 (33%), Positives = 22/39 (56%), Gaps = 4/39 (10%)
Query 1 RPKLNLFDLSSFISFLLLQD----FSKPRKMENDEGRVV 35
RPK+ LFD+ S F + FS+P+K+E+ + +
Sbjct 1465 RPKMTLFDMQSLEIFFQIAACEALFSEPKKLESVQSNIT 1503
> Hs7705308
Length=429
Score = 26.9 bits (58), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 28/62 (45%), Gaps = 4/62 (6%)
Query 50 IPAKEHGAVQINVGMVDEAGRYTGEYTTVAISGAVRQRGESDA----CMNRLMFEKKILS 105
+P+++ V+ M+D RYT + +T S VR DA F+K IL
Sbjct 75 VPSQDKTRVEPPQYMIDLYNRYTSDKSTTPASNIVRSFSMEDAISITATEDFPFQKHILL 134
Query 106 FS 107
F+
Sbjct 135 FN 136
Lambda K H
0.320 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160781780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40