bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3725_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
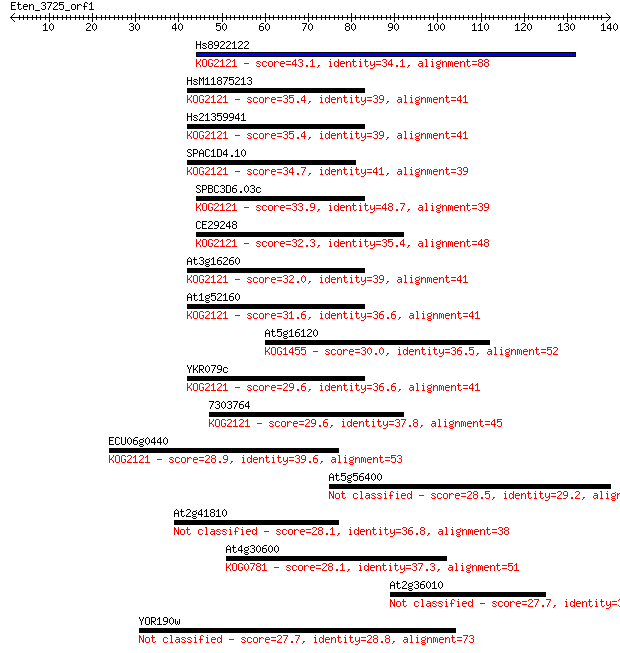
Hs8922122 43.1 1e-04
HsM11875213 35.4 0.031
Hs21359941 35.4 0.031
SPAC1D4.10 34.7 0.064
SPBC3D6.03c 33.9 0.088
CE29248 32.3 0.27
At3g16260 32.0 0.34
At1g52160 31.6 0.43
At5g16120 30.0 1.4
YKR079c 29.6 1.8
7303764 29.6 1.9
ECU06g0440 28.9 2.7
At5g56400 28.5 4.0
At2g41810 28.1 4.8
At4g30600 28.1 5.2
At2g36010 27.7 7.7
YOR190w 27.7 7.8
> Hs8922122
Length=363
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 46/90 (51%), Gaps = 15/90 (16%)
Query 44 AFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFLRSVLT--MERVEEEARQVYDI 101
A E GHS+ MA +FA A RL+LTHFSQRY+ R T + ++++A V D+
Sbjct 286 AKEHGHSTPQMAATFAKLCRAKRLVLTHFSQRYKPVALAREGETDGIAELKKQAESVLDL 345
Query 102 ERQLQAASLPASPLKVTAAWDGLAVVLPQR 131
+ +VT A D + + +P +
Sbjct 346 Q-------------EVTLAEDFMVISIPIK 362
> HsM11875213
Length=826
Score = 35.4 bits (80), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
+ A E+ HS+ + A S R A ++L HFSQRY P
Sbjct 695 EEAVEKTHSTTSQAISVGMRMNAEFIMLNHFSQRYAKVPLF 735
> Hs21359941
Length=826
Score = 35.4 bits (80), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
+ A E+ HS+ + A S R A ++L HFSQRY P
Sbjct 695 EEAVEKTHSTTSQAISVGMRMNAEFIMLNHFSQRYAKVPLF 735
> SPAC1D4.10
Length=809
Score = 34.7 bits (78), Expect = 0.064, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDP 80
+ A ++ HS+ + A A +AG ++LTHFSQRY P
Sbjct 718 EIAIKKQHSTYSEALEVAKKAGTKNVILTHFSQRYPKLP 756
> SPBC3D6.03c
Length=678
Score = 33.9 bits (76), Expect = 0.088, Method: Composition-based stats.
Identities = 19/39 (48%), Positives = 23/39 (58%), Gaps = 0/39 (0%)
Query 44 AFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
A +R HS+A+ A S A A L+LTHFSQR FL
Sbjct 608 AIQRQHSTASEALSVAQSMKAKALILTHFSQRSYDADFL 646
> CE29248
Length=850
Score = 32.3 bits (72), Expect = 0.27, Method: Composition-based stats.
Identities = 17/48 (35%), Positives = 22/48 (45%), Gaps = 0/48 (0%)
Query 44 AFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFLRSVLTMERV 91
A + HS+ A R A ++LTHFS RY P L L E +
Sbjct 745 AMRKRHSTMGQAVDVGKRMNAKHIILTHFSARYPKVPVLPEYLDKENI 792
> At3g16260
Length=937
Score = 32.0 bits (71), Expect = 0.34, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
+ A + HS+ A AG R +LTHFSQRY P +
Sbjct 831 EEAVAKNHSTTKEAIKVGSSAGVYRTVLTHFSQRYPKIPVI 871
> At1g52160
Length=837
Score = 31.6 bits (70), Expect = 0.43, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
+ A + HS+ A A R++LTHFSQRY P +
Sbjct 738 EEALAKNHSTTKEAIDVGSAANVYRIVLTHFSQRYPKIPVI 778
> At5g16120
Length=340
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 25/52 (48%), Gaps = 6/52 (11%)
Query 60 FRAGAARLLLTHFSQRYRGDPFLRSVLTMERVEEEARQVYDIERQLQAASLP 111
FR R + + Y G P LR+ + M R + DIE+QLQ SLP
Sbjct 219 FRDIRKRDMTPYNMICYSGKPRLRTAVEMLRTTQ------DIEKQLQEVSLP 264
> YKR079c
Length=838
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 15/41 (36%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 42 QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFL 82
+ A ++ H + A + + A +L+LTHFSQRY P L
Sbjct 730 EDAVKKKHCTINEAIGVSNKMNARKLILTHFSQRYPKLPQL 770
> 7303764
Length=743
Score = 29.6 bits (65), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 47 RGHSSAAMAGSFAFRAGAARLLLTHFSQRYRGDPFLRSVLTMERV 91
+ HS+ + A A +L+HFSQRY P L S M+RV
Sbjct 644 KTHSTVSQAIQQGRNMNARHTILSHFSQRYAKCPRLPSDEDMQRV 688
> ECU06g0440
Length=744
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 26/56 (46%), Gaps = 3/56 (5%)
Query 24 GKALSAKDLMGAHVQTAS---QAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRY 76
G S D+M AS + A GHS+ A + + LLLTHFSQRY
Sbjct 645 GMMSSNSDVMIHEATFASDQQERAAHTGHSTIDEAAKIFEMSQSRTLLLTHFSQRY 700
> At5g56400
Length=450
Score = 28.5 bits (62), Expect = 4.0, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 32/75 (42%), Gaps = 10/75 (13%)
Query 75 RYRGD----------PFLRSVLTMERVEEEARQVYDIERQLQAASLPASPLKVTAAWDGL 124
+Y GD P LR+++ E+ ++ I R LQ+ ++ SPL A D
Sbjct 192 KYSGDESVRTLISSCPSLRNLVVKRHNEDNVKRFAIIVRYLQSLTVYLSPLHGVADSDAY 251
Query 125 AVVLPQRAYSPTEEH 139
+ P Y ++H
Sbjct 252 VINTPNLKYLNIKDH 266
> At2g41810
Length=370
Score = 28.1 bits (61), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 39 TASQAAFERGHSSAAMAGSFAFRAGAARLLLTHFSQRY 76
+A + FE A G FAFRA + R +T +S Y
Sbjct 309 SAFKVTFESNDKGAFKVGRFAFRADSNRTRITFYSGFY 346
> At4g30600
Length=634
Score = 28.1 bits (61), Expect = 5.2, Method: Composition-based stats.
Identities = 19/59 (32%), Positives = 26/59 (44%), Gaps = 11/59 (18%)
Query 51 SAAMAGSFAFRAGAARLLLTH--------FSQRYRGDPFLRSVLTMERVEEEARQVYDI 101
S MA FR+GA L TH F + Y DP +V+ E ++E R D+
Sbjct 458 SVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDP---AVVAKEAIQEATRNGSDV 513
> At2g36010
Length=532
Score = 27.7 bits (60), Expect = 7.7, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 18/36 (50%), Gaps = 0/36 (0%)
Query 89 ERVEEEARQVYDIERQLQAASLPASPLKVTAAWDGL 124
E +E + R++YDI L+ L P K W G+
Sbjct 262 ETLEVQKRRIYDITNVLEGIDLIEKPFKNRILWKGV 297
> YOR190w
Length=445
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 21/77 (27%), Positives = 34/77 (44%), Gaps = 11/77 (14%)
Query 31 DLMGAHVQTASQAAFERGHSSAAMAGSFAF----RAGAARLLLTHFSQRYRGDPFLRSVL 86
DL GA A + G ++ + S+ F A LT+ +Y D +L +V+
Sbjct 174 DLHGA-------AGSQNGFDNSGLRDSYKFLEDENLSATMKALTYILSKYSTDVYLDTVI 226
Query 87 TMERVEEEARQVYDIER 103
+E + E V D+ER
Sbjct 227 GIELLNEPLGPVIDMER 243
Lambda K H
0.317 0.128 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40