bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
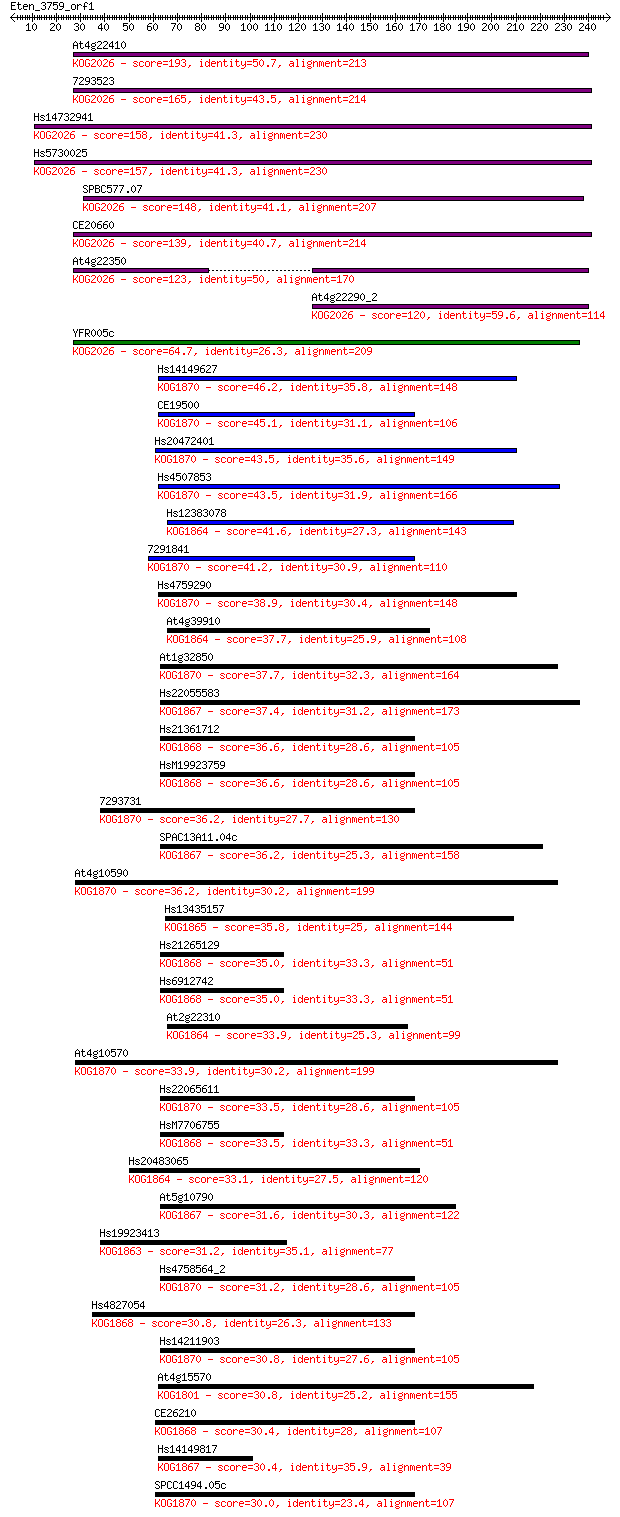
Query= Eten_3759_orf1
Length=248
Score E
Sequences producing significant alignments: (Bits) Value
At4g22410 193 3e-49
7293523 165 6e-41
Hs14732941 158 9e-39
Hs5730025 157 1e-38
SPBC577.07 148 1e-35
CE20660 139 4e-33
At4g22350 123 3e-28
At4g22290_2 120 2e-27
YFR005c 64.7 2e-10
Hs14149627 46.2 6e-05
CE19500 45.1 1e-04
Hs20472401 43.5 4e-04
Hs4507853 43.5 4e-04
Hs12383078 41.6 0.002
7291841 41.2 0.002
Hs4759290 38.9 0.009
At4g39910 37.7 0.019
At1g32850 37.7 0.022
Hs22055583 37.4 0.026
Hs21361712 36.6 0.048
HsM19923759 36.6 0.048
7293731 36.2 0.059
SPAC13A11.04c 36.2 0.065
At4g10590 36.2 0.069
Hs13435157 35.8 0.075
Hs21265129 35.0 0.14
Hs6912742 35.0 0.15
At2g22310 33.9 0.29
At4g10570 33.9 0.32
Hs22065611 33.5 0.39
HsM7706755 33.5 0.42
Hs20483065 33.1 0.57
At5g10790 31.6 1.6
Hs19923413 31.2 1.9
Hs4758564_2 31.2 2.2
Hs4827054 30.8 2.4
Hs14211903 30.8 3.0
At4g15570 30.8 3.0
CE26210 30.4 3.8
Hs14149817 30.4 3.9
SPCC1494.05c 30.0 5.1
> At4g22410
Length=340
Score = 193 bits (491), Expect = 3e-49, Method: Compositional matrix adjust.
Identities = 108/217 (49%), Positives = 146/217 (67%), Gaps = 5/217 (2%)
Query 27 DTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTI 86
D ++ LNP ++ V L + ++LDG+D+LPG VGLNN+ KT+F V IQSL +
Sbjct 78 DDIRHVLNPRFSRAQVNELDKNRQWSRALDGSDYLPGMVGLNNIQKTEFVNVTIQSLMRV 137
Query 87 IPLRNYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFR 146
PLRN+ + + + K + ++ EL RKI++ RNFKG VSPHEFLQAV SKK+FR
Sbjct 138 TPLRNFFHIPENYQHCK-SPLVHCFGELTRKIWHARNFKGQVSPHEFLQAVMKASKKRFR 196
Query 147 IGEQSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGEV-LVTDAQSEEGR---KTPFL 202
IG+QSDP+ +WLLN LH D + K SIIH CFQGE+ +V + Q E + + FL
Sbjct 197 IGQQSDPVEFMSWLLNTLHMDLRTSKDASSIIHKCFQGELEVVKEFQGNENKEISRMSFL 256
Query 203 YLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKFDGET 239
L LDLPP P+FKD +++N+IP V +FDLL+KFDGET
Sbjct 257 MLGLDLPPPPLFKDVMEKNIIPQVALFDLLKKFDGET 293
> 7293523
Length=719
Score = 165 bits (418), Expect = 6e-41, Method: Compositional matrix adjust.
Identities = 93/247 (37%), Positives = 136/247 (55%), Gaps = 38/247 (15%)
Query 27 DTFKYNLNPTYTPEDVARLSA-EVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCT 85
D KY LNPT+T +++++L + + +++DG +LPG VGLNN+ D+ V++ +L
Sbjct 109 DDIKYVLNPTFTRQEISKLDQLQPKHSRTVDGVLYLPGVVGLNNIKANDYCNVVLHALSH 168
Query 86 IIPLRNYLLLLDLSEKQKYNSVLK-----------TLSELMRKIFNRRNFKGIVSPHEFL 134
+ PLR+Y L +KQ Y V++ ELMRK++N RNFK VSPHE L
Sbjct 169 VGPLRDYFL-----QKQSYAHVVRPPGDSVFTLVQRFGELMRKMWNPRNFKSHVSPHEML 223
Query 135 QAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQK-DKKTGGSIIHDCFQGEV------- 186
QAV + S K+F+I EQ DP+ +W LN LHR K +K SI++ F GE+
Sbjct 224 QAVVLWSSKRFQITEQGDPIDFLSWFLNTLHRALKGNKHPNSSILYKIFLGEMKIYTRKM 283
Query 187 -------------LVTDAQSEEGRKTPFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQ 233
L T+ ++ F+YLT DLPP P+F D N+IP V ++ LL
Sbjct 284 PPVELDDAAKAQLLATEEYKDQVEDKNFIYLTCDLPPPPLFTDEFRENIIPQVNLYQLLS 343
Query 234 KFDGETD 240
KF+G +
Sbjct 344 KFNGTAE 350
> Hs14732941
Length=404
Score = 158 bits (400), Expect = 9e-39, Method: Compositional matrix adjust.
Identities = 95/262 (36%), Positives = 138/262 (52%), Gaps = 40/262 (15%)
Query 11 AFAFPTSTSTPVWFAGDTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNL 70
AF +S P W L PT+T + +A L + ++ DG +LPG VGLNN+
Sbjct 13 AFQTTMRSSIPHWRISRCV---LKPTFTKQQIANLDKQAKLSRAYDGTTYLPGIVGLNNI 69
Query 71 SKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSV-----------LKTLSELMRKIF 119
D+ ++Q+L + PLRNY L E+ Y ++ ++ ELMRK++
Sbjct 70 KANDYANAVLQALSNVPPLRNYFL-----EEDNYKNIKRPPGDIMFLLVQRFGELMRKLW 124
Query 120 NRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD-QKDKKTGGSII 178
N RNFK VSPHE LQAV + SKK F+I +Q D + +W LN LH KK +I+
Sbjct 125 NPRNFKAHVSPHEMLQAVVLCSKKTFQITKQGDGVDFLSWFLNALHSALGGTKKKKKTIV 184
Query 179 HDCFQG--------------------EVLVTDAQSEEGRKTPFLYLTLDLPPAPIFKDSL 218
D FQG ++L D E ++ F+YLTLDLP AP++KD
Sbjct 185 TDVFQGSMRIFTKKLPHPDLPAEEKEQLLHNDEYQETMVESTFMYLTLDLPTAPLYKDEK 244
Query 219 DRNMIPTVPIFDLLQKFDGETD 240
++ +IP VP+F++L KF+G T+
Sbjct 245 EQLIIPQVPLFNILAKFNGITE 266
> Hs5730025
Length=324
Score = 157 bits (398), Expect = 1e-38, Method: Compositional matrix adjust.
Identities = 95/262 (36%), Positives = 139/262 (53%), Gaps = 40/262 (15%)
Query 11 AFAFPTSTSTPVWFAGDTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNL 70
AF +S P W + L PT+T + +A L + ++ DG +LPG VGLNN+
Sbjct 13 AFQTTMRSSIPHWRIS---RMCLKPTFTKQQIANLDKQAKLSRAYDGTTYLPGIVGLNNI 69
Query 71 SKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSV-----------LKTLSELMRKIF 119
D+ ++Q+L + PLRNY L E+ Y ++ ++ ELMRK++
Sbjct 70 KANDYANAVLQALSNVPPLRNYFL-----EEDNYKNIKRPPGDIMFLLVQRFGELMRKLW 124
Query 120 NRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD-QKDKKTGGSII 178
N RNFK VSPHE LQAV + SKK F+I +Q D + +W LN LH KK +I+
Sbjct 125 NPRNFKAHVSPHEMLQAVVLCSKKTFQITKQGDGVDFLSWFLNALHSALGGTKKKKKTIV 184
Query 179 HDCFQG--------------------EVLVTDAQSEEGRKTPFLYLTLDLPPAPIFKDSL 218
D FQG ++L D E ++ F+YLTLDLP AP++KD
Sbjct 185 TDVFQGSMRIFTKKLPHPDLPAEEKEQLLHNDEYQETMVESTFMYLTLDLPTAPLYKDEK 244
Query 219 DRNMIPTVPIFDLLQKFDGETD 240
++ +IP VP+F++L KF+G T+
Sbjct 245 EQLIIPQVPLFNILAKFNGITE 266
> SPBC577.07
Length=502
Score = 148 bits (373), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 85/228 (37%), Positives = 123/228 (53%), Gaps = 24/228 (10%)
Query 31 YNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLR 90
Y + PT+T +V RL L ++PG VG+NN+ D+F V+I L + P R
Sbjct 144 YVMRPTFTKLEVQRLDHTPQLSYDLMLKPYVPGFVGMNNIKNNDYFNVVIHMLAHVKPFR 203
Query 91 NYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQ 150
NY LL + + +++ L+ L+RK++N + FK VSP E +Q V V S KK+ I EQ
Sbjct 204 NYFLLKNFDNCPQ---LVQRLAILIRKLWNHKAFKSHVSPQELIQEVTVLSHKKYSINEQ 260
Query 151 SDPLALFAWLLNVLHRDQKDKKTG----GSIIHDCFQGEVLVTD----AQSEEGRK---- 198
DP+ +W LN LH KK+ SI+H FQG V + +E+G +
Sbjct 261 KDPVEFLSWFLNTLHNCLGGKKSTIAKPTSIVHYSFQGFVRIESQKIRQHAEKGEQVVFT 320
Query 199 ---------TPFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKFDG 237
PFLYLTLDLPP PIF+D + N+IP V + ++L K++G
Sbjct 321 GDRVIQTNVVPFLYLTLDLPPKPIFQDEFEGNIIPQVELKEILNKYNG 368
> CE20660
Length=602
Score = 139 bits (351), Expect = 4e-33, Method: Compositional matrix adjust.
Identities = 87/245 (35%), Positives = 126/245 (51%), Gaps = 36/245 (14%)
Query 27 DTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTI 86
+ KY L PTYT E +A + + ++ D + + PG VGLNN+ D+ I+Q+L +
Sbjct 218 EDIKYVLKPTYTKEMIASIDKQSKMVRAYDDSTYFPGVVGLNNIKANDYCNAILQALSAV 277
Query 87 IPLRNYLLLLDLSEKQKYNSVLK-----------TLSELMRKIFNRRNFKGIVSPHEFLQ 135
PLRN+ L ++ Y S+ + EL+RK++N R + VSPHE LQ
Sbjct 278 RPLRNWFL-----KESNYTSIKRPPGDKLTLLPQRFGELIRKLWNPRALRTHVSPHEMLQ 332
Query 136 AVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGE--------VL 187
AV V S KKF+ +Q+D +LL LH SII F+G +
Sbjct 333 AVVVCSNKKFQFIKQNDAADFMLFLLTTLHSALNGTDKKPSIISKTFRGRMRQYSRRVIP 392
Query 188 VTDAQSEEGRK------------TPFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKF 235
D E+ K +PFLYLTLDLP AP+++D +N+IP VP+ LL+KF
Sbjct 393 AEDTDEEKYMKLRMPEYQEKVIESPFLYLTLDLPSAPLYRDVQLQNIIPQVPLSTLLEKF 452
Query 236 DGETD 240
DG+T+
Sbjct 453 DGKTE 457
> At4g22350
Length=477
Score = 123 bits (309), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 69/118 (58%), Positives = 87/118 (73%), Gaps = 4/118 (3%)
Query 126 GIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGE 185
G+VSPHEFLQAV SKK+FRIG+QSDP+ +WLLN LH D + K SIIH CFQGE
Sbjct 227 GMVSPHEFLQAVMKASKKRFRIGQQSDPVEFMSWLLNTLHMDLRTSKDASSIIHKCFQGE 286
Query 186 V-LVTDAQSEEGR---KTPFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKFDGET 239
+ +V + Q E + + PFL L LDLPP P+FKD +++N+IP V +FDLL+KFDGET
Sbjct 287 LEVVKEYQGNENKEISRMPFLMLGLDLPPPPLFKDVMEKNIIPQVALFDLLKKFDGET 344
Score = 34.3 bits (77), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 16/56 (28%), Positives = 30/56 (53%), Gaps = 5/56 (8%)
Query 27 DTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQS 82
D ++ LNP ++ V L + ++LDG+D+LPG V S +F ++++
Sbjct 191 DDIRHVLNPRFSRAQVNELDKNRQWSRALDGSDYLPGMV-----SPHEFLQAVMKA 241
> At4g22290_2
Length=543
Score = 120 bits (302), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 68/118 (57%), Positives = 86/118 (72%), Gaps = 4/118 (3%)
Query 126 GIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGE 185
G+VSPHEFLQAV SKK+FRIG+QSDP+ +WLLN LH D + K SIIH CFQGE
Sbjct 282 GMVSPHEFLQAVMKASKKRFRIGQQSDPVEFMSWLLNTLHMDLRTSKDASSIIHKCFQGE 341
Query 186 V-LVTDAQSEEGR---KTPFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKFDGET 239
+ +V + Q E + + FL L LDLPP P+FKD +++N+IP V +FDLL+KFDGET
Sbjct 342 LEVVKEFQGNENKEISRMSFLMLGLDLPPPPLFKDVMEKNIIPQVALFDLLKKFDGET 399
> YFR005c
Length=448
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 55/218 (25%), Positives = 101/218 (46%), Gaps = 21/218 (9%)
Query 27 DTFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTI 86
++ K+ PTY P+D+ + F L +L G +G N + D+ ++ + +
Sbjct 115 NSIKFAAYPTYCPKDLEDFPRQCF---DLSNRTYLNGFIGFTNAATYDYAHSVLLLISHM 171
Query 87 IPLRNYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFR 146
+P+R++ LL + ++ +K LS ++KI++ + FK +S +F+ + V
Sbjct 172 VPVRDHFLLNHFDNQGEF---IKRLSICVKKIWSPKLFKHHLSVDDFVSYLKVREGLNLN 228
Query 147 IGEQSDPLALFAWLLNVLHRDQKDKKTGGSIIHDCFQGEVLVTDAQSE-EGRKT------ 199
DP WL N + D K SI++ +G+V + +++ E ++
Sbjct 229 ---PIDPRLFLLWLFNKICSSSNDLK---SILNHSCKGKVKIAKVENKPEASESVTGKVI 282
Query 200 --PFLYLTLDLPPAPIFKDSLDRNMIPTVPIFDLLQKF 235
PF LTLDLP F+D + +P + I LL KF
Sbjct 283 VKPFWVLTLDLPEFSPFEDGNSVDDLPQINITKLLTKF 320
> Hs14149627
Length=952
Score = 46.2 bits (108), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 53/187 (28%), Positives = 76/187 (40%), Gaps = 42/187 (22%)
Query 62 PGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNS-------VLKTLSEL 114
PG GL+NL T F IQ L PL Y L E+ +++ + K+ +EL
Sbjct 257 PGLCGLSNLGNTCFMNSAIQCLSNTPPLTEYFLNDKYQEELNFDNPLGMRGEIAKSYAEL 316
Query 115 MRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD------- 167
++++++ + V+P F VG +F +Q D L A+LL+ LH D
Sbjct 317 IKQMWSGK--FSYVTPRAFKTQVG-RFAPQFSGYQQQDCQELLAFLLDGLHEDLNRIRKK 373
Query 168 ----------QKDK-----------KTGGSIIHDCFQGEVLVTDAQSEEGRKT----PFL 202
+ DK K SII D F G T E + + PF
Sbjct 374 PYIQLKDADGRPDKVVAEEAWENHLKRNDSIIVDIFHGLFKSTLVCPECAKISVTFDPFC 433
Query 203 YLTLDLP 209
YLTL LP
Sbjct 434 YLTLPLP 440
> CE19500
Length=900
Score = 45.1 bits (105), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/113 (29%), Positives = 53/113 (46%), Gaps = 10/113 (8%)
Query 62 PGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL----LDLSEKQ---KYNSVLKTLSEL 114
PG GL+NL T F A Q L + PLR Y L D++E + + + EL
Sbjct 272 PGACGLSNLGNTCFMASAFQCLSNMPPLREYFLANTYQNDINEDNPLGTHGHLAMAVGEL 331
Query 115 MRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
M+ +++ ++P +F +G + +F Q D L A++L+ LH D
Sbjct 332 MKGMWS--GDYASINPRKFKSIIG-QFAPRFNGYSQQDAHELMAYVLDGLHED 381
> Hs20472401
Length=1320
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 53/187 (28%), Positives = 71/187 (37%), Gaps = 40/187 (21%)
Query 61 LPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSELMR--KI 118
LPG GL NL T F +IQSL LR++ + YN+ L T L +
Sbjct 495 LPGFTGLVNLGNTCFMNSVIQSLSNTRELRDFFHDRSFEAEINYNNPLGTGGRLAIGFAV 554
Query 119 FNRRNFKGIVSPHEF----LQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD------- 167
R +KG + H F L+A+ +F Q D A+LL+ LH D
Sbjct 555 LLRALWKG--THHAFQPSKLKAIVASKASQFTGYAQHDAQEFMAFLLDGLHEDLNRIQNK 612
Query 168 ---------------------QKDKKTGGSIIHDCFQGEV---LVTDAQSEEGRK-TPFL 202
Q+ K S I D FQG+ LV ++ PFL
Sbjct 613 PYTETVDSDGRPDEVVAEEAWQRHKMRNDSFIVDLFQGQYKSKLVCPVCAKVSITFDPFL 672
Query 203 YLTLDLP 209
YL + LP
Sbjct 673 YLPVPLP 679
> Hs4507853
Length=963
Score = 43.5 bits (101), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 53/206 (25%), Positives = 83/206 (40%), Gaps = 46/206 (22%)
Query 62 PGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDL-SEKQKYN------SVLKTLSEL 114
PG GL NL T F +Q L PL +Y L + +E + N + + +EL
Sbjct 299 PGLCGLGNLGNTCFMNSALQCLSNTAPLTDYFLKDEYEAEINRDNPLGMKGEIAEAYAEL 358
Query 115 MRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD------- 167
++++++ R+ V+P F VG +F +Q D L A+LL+ LH D
Sbjct 359 IKQMWSGRD--AHVAPSMFKTQVG-RFAPQFSGYQQQDSQELLAFLLDGLHEDLNRVKKK 415
Query 168 ---------------------QKDKKTGGSIIHDCFQGEVLVTDAQSEEGRKT----PFL 202
+ + S+I D F G T E + + PF
Sbjct 416 PYLELKDANGRPDAVVAKEAWENHRLRNDSVIVDTFHGLFKSTLVCPECAKVSVTFDPFC 475
Query 203 YLTLDLPPAPIFKDS-LDRNMIPTVP 227
YLTL P P+ KD ++ ++P P
Sbjct 476 YLTL---PLPLKKDRVMEVFLVPADP 498
> Hs12383078
Length=366
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 39/169 (23%), Positives = 72/169 (42%), Gaps = 28/169 (16%)
Query 66 GLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFK 125
GL N T + ++Q+L P R +L +K+K N +L L++L I ++
Sbjct 36 GLVNFGNTCYCNSVLQALYFCRPFRENVLAYKAQQKKKEN-LLTCLADLFHSIATQKKKV 94
Query 126 GIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLN----VLHRDQKDKKTGGSI---- 177
G++ P +F+ + E+ F Q D +LLN +L ++K +K G +
Sbjct 95 GVIPPKKFISRLRKEN-DLFDNYMQQDAHEFLNYLLNTIADILQEEKKQEKQNGKLKNGN 153
Query 178 --------------IHDCFQG----EVLVTDAQSEEGRKTPFLYLTLDL 208
+H+ FQG E + ++ + FL L++D+
Sbjct 154 MNEPAENNKPELTWVHEIFQGTLTNETRCLNCETVSSKDEDFLDLSVDV 202
> 7291841
Length=1096
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 34/123 (27%), Positives = 52/123 (42%), Gaps = 17/123 (13%)
Query 58 ADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL----LDLSEKQKYNS------- 106
AD PG +GL N T F ++Q L L Y +L DL + K NS
Sbjct 62 ADQTPGVMGLKNHGNTCFMNAVLQCLSHTDILAEYFVLDQYKADLKRRNKINSRKFGTKG 121
Query 107 -VLKTLSELMRKIFNRRNFKGIVSPHEF-LQAVGVESKKKFRIGEQSDPLALFAWLLNVL 164
+ + L+ +++ ++ +N S H +AV +FR Q D WLL+ +
Sbjct 122 ELTEQLANVLKALWTCKN----ESDHSTSFKAVVDRYGTQFRSSTQHDAQEFLFWLLDKV 177
Query 165 HRD 167
H D
Sbjct 178 HED 180
> Hs4759290
Length=690
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 45/187 (24%), Positives = 73/187 (39%), Gaps = 42/187 (22%)
Query 62 PGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNS-------VLKTLSEL 114
PG GL NL T F +Q L + L Y L E+ + + + + ++L
Sbjct 33 PGICGLTNLGNTCFMNSALQCLSNVPQLTEYFLNNCYLEELNFRNPLGMKGEIAEAYADL 92
Query 115 MRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD------- 167
+++ ++ + + IV PH F VG +F +Q D L ++LL+ LH D
Sbjct 93 VKQAWSGHH-RSIV-PHVFKNKVG-HFASQFLGYQQHDSQELLSFLLDGLHEDLNRVKKK 149
Query 168 ---------------------QKDKKTGGSIIHDCFQG----EVLVTDAQSEEGRKTPFL 202
Q K+ S+I D F G ++ D + PF
Sbjct 150 EYVELCDAAGRPDQEVAQEAWQNHKRRNDSVIVDTFHGLFKSTLVCPDCGNVSVTFDPFC 209
Query 203 YLTLDLP 209
YL++ LP
Sbjct 210 YLSVPLP 216
> At4g39910
Length=371
Score = 37.7 bits (86), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 28/115 (24%), Positives = 55/115 (47%), Gaps = 8/115 (6%)
Query 66 GLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNS---VLKTLSELMRKIFNRR 122
G N T + ++Q+L +P R LL S K ++ ++ L++L +I +++
Sbjct 24 GFENFGNTCYCNSVLQALYFCVPFREQLLEYYTSNKSVADAEENLMTCLADLFSQISSQK 83
Query 123 NFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLN----VLHRDQKDKKT 173
G+++P F+Q + ++ + FR D +LLN +L ++ K KT
Sbjct 84 KKTGVIAPKRFVQRLKKQN-ELFRSYMHQDAHEFLNYLLNEVVDILEKEAKATKT 137
> At1g32850
Length=887
Score = 37.7 bits (86), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 53/202 (26%), Positives = 77/202 (38%), Gaps = 47/202 (23%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYN------SVLKTLSELMR 116
G GL NL T F +Q L P+ Y L S+ N + EL+R
Sbjct 294 GLGGLQNLGNTCFMNSTLQCLAHTPPIVEYFLQDYRSDINAKNPLGMRGELAIAFGELLR 353
Query 117 KIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD--------- 167
K+++ + + V+P F + +F Q D + A+LL+ LH D
Sbjct 354 KLWS--SGQNTVAPRAFKTKLA-RFAPQFSGYNQHDSQEMLAFLLDGLHEDLNKVKRKPY 410
Query 168 --QKD-----------------KKTGGSIIHDCFQGEVLVTDAQSEEGRKT----PFLYL 204
KD K S+I D FQG+ T + G+ + PF+YL
Sbjct 411 IEAKDSDGRPDDEVAEEKWKYHKARNDSVIVDVFQGQYKSTLVCPDCGKISITFDPFMYL 470
Query 205 TLDLPPAPIFKDSLDRNMIPTV 226
+L LP S R+M TV
Sbjct 471 SLPLPS------SRTRSMTVTV 486
> Hs22055583
Length=800
Score = 37.4 bits (85), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 54/204 (26%), Positives = 82/204 (40%), Gaps = 39/204 (19%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSEL--MRKIFN 120
G GL NL T F I+Q+L I L+++ L S+K K +L + M +F+
Sbjct 450 GLRGLINLGNTCFMNCIVQALTHIPLLKDFFL----SDKHKCIMTSPSLCLVCEMSSLFH 505
Query 121 RRNFKGIVSPH---EFLQAVGVESKK--KFRIGEQSDPLALFAWLLNVLHRDQKDKKTG- 174
+ G +PH + L + + ++ +R Q D +L+VLHR KD G
Sbjct 506 AM-YSGSRTPHIPYKLLHLIWIHAEHLAGYR---QQDAHEFLIAILDVLHRHSKDDSGGQ 561
Query 175 --------GSIIHDCFQG----EVLVTDAQSEEGRKTPFLYLTLDLPPAPIFKDSLD--- 219
II F G +V S P ++LDLP + DS +
Sbjct 562 EANNPNCCNCIIDQIFTGGLQSDVTCQACHSVSTTIDPCWDISLDLPGSCATFDSQNPER 621
Query 220 -------RNMIPTVP-IFDLLQKF 235
+ IP +P + D LQ F
Sbjct 622 ADSTVSRDDHIPGIPSLTDCLQWF 645
> Hs21361712
Length=605
Score = 36.6 bits (83), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 50/110 (45%), Gaps = 7/110 (6%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLL----LLDLSE-KQKYNSVLKTLSELMRK 117
G GL NL T F I+Q L LR+Y L + DL + ++++ ++L++
Sbjct 265 GLAGLRNLGNTCFMNSILQCLSNTRELRDYCLQRLYMRDLHHGSNAHTALVEEFAKLIQT 324
Query 118 IFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
I+ + +VSP EF + +F Q D +LL+ LH +
Sbjct 325 IWT-SSPNDVVSPSEFKTQI-QRYAPRFVGYNQQDAQEFLRFLLDGLHNE 372
> HsM19923759
Length=605
Score = 36.6 bits (83), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 30/110 (27%), Positives = 50/110 (45%), Gaps = 7/110 (6%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLL----LLDLSE-KQKYNSVLKTLSELMRK 117
G GL NL T F I+Q L LR+Y L + DL + ++++ ++L++
Sbjct 265 GLAGLRNLGNTCFMNSILQCLSNTRELRDYCLQRLYMRDLHHGSNAHTALVEEFAKLIQT 324
Query 118 IFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
I+ + +VSP EF + +F Q D +LL+ LH +
Sbjct 325 IWT-SSPNDVVSPSEFKTQI-QRYAPRFVGYNQQDAQEFLRFLLDGLHNE 372
> 7293731
Length=1351
Score = 36.2 bits (82), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 16/137 (11%)
Query 38 TPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNY----L 93
TP D RL+ +S + PG GL+NL T F +Q L PL Y +
Sbjct 699 TPGDRRRLT------RSSIMSVHAPGATGLHNLGNTCFMNAALQVLFNTQPLAQYFQREM 752
Query 94 LLLDLSEKQKYNS---VLKTLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQ 150
+++ K + + +EL+++++ V+P + V + +F G Q
Sbjct 753 HRFEVNAANKLGTKGQLAMRYAELLKEVWTATTRS--VAPLKLRFCVN-KYAPQFAGGGQ 809
Query 151 SDPLALFAWLLNVLHRD 167
D L WLL+ LH D
Sbjct 810 HDSQELLEWLLDALHED 826
> SPAC13A11.04c
Length=449
Score = 36.2 bits (82), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 40/171 (23%), Positives = 69/171 (40%), Gaps = 20/171 (11%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLL--LLDLSEKQKYNSVLKTLSELMRKIFN 120
G G+ NL T F +VI+QS+ +RN ++ ++ + + ++ I+N
Sbjct 143 GLRGIQNLGATCFMSVILQSILHNPLVRNLFFSGFHTSTDCKRPTCMTCAIDDMFSSIYN 202
Query 121 RRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQKDKKTGGSI--- 177
+N P L + SK +Q D F +LL+ +H + +GG
Sbjct 203 SKNKSTFYGPTAVLNLMWKLSKSLCGYSQQ-DGHEFFVYLLDQMHTE-----SGGGTSMP 256
Query 178 ----IHDCFQGE----VLVTDAQSEEGRKTPFLYLTLDLPPAPIFKDSLDR 220
IH F G V D + E P + ++LD+ P + L+R
Sbjct 257 CTCPIHRIFSGSLKNVVTCLDCKKERVAVDPLMDISLDINE-PTLQGCLER 306
> At4g10590
Length=937
Score = 36.2 bits (82), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 60/238 (25%), Positives = 90/238 (37%), Gaps = 55/238 (23%)
Query 28 TFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTII 87
T ++L P T ED S + GK G G GL+NL T F +Q L
Sbjct 299 TSNFSLFPRITSEDDGSNSLSIL-GKGEKG-----GLAGLSNLGNTCFMNSALQCLAHTP 352
Query 88 PLRNYLLLLDLSEKQKYNSVLKTLSE-------LMRKIFNRRNFKGIVSPHEFLQAVGVE 140
P+ Y L D S+ ++ L E L++K+++ + + V+P F +
Sbjct 353 PIVEY-FLQDYSDDINRDNPLGMCGELAIAFGDLLKKLWS--SGRNSVAPRAFKTKLA-R 408
Query 141 SKKKFRIGEQSDPLALFAWLLNVLHRD----------------------------QKDKK 172
+F Q D L A+LL+ LH D K
Sbjct 409 FAPQFSGYNQHDSQELLAFLLDGLHEDLNKVKRKPYIELKDSDSRPDDEVAEELWNYHKA 468
Query 173 TGGSIIHDCFQGEVLVTDAQSEEGRKT----PFLYLTLDLPPAPIFKDSLDRNMIPTV 226
S+I D QG+ T G+ + PF+YL++ LP +L R+M TV
Sbjct 469 RNDSVIVDVCQGQYKSTLVCPACGKISITFDPFMYLSVPLP------STLTRSMTVTV 520
> Hs13435157
Length=548
Score = 35.8 bits (81), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 36/157 (22%), Positives = 66/157 (42%), Gaps = 16/157 (10%)
Query 65 VGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNS--VLKTLSELMRKIFNRR 122
GL+NL T F IQ L PL NYLL + + S +L + + + F
Sbjct 122 AGLHNLGNTCFLNATIQCLTYTPPLANYLLSKEHARSCHQGSFCMLCVMQNHIVQAF--A 179
Query 123 NFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHR------DQKDKKTGG- 175
N + P F++ + + + FR G Q D + ++ + + + D++T
Sbjct 180 NSGNAIKPVSFIRDLK-KIARHFRFGNQEDAHEFLRYTIDAMQKACLNGCAKLDRQTQAT 238
Query 176 SIIHDCFQG----EVLVTDAQSEEGRKTPFLYLTLDL 208
+++H F G V + +S P+L + L++
Sbjct 239 TLVHQIFGGYLRSRVKCSVCKSVSDTYDPYLDVALEI 275
> Hs21265129
Length=551
Score = 35.0 bits (79), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSE 113
G VGL NL T F ++Q L + PLR++ L D ++ + L+E
Sbjct 210 GHVGLRNLGNTCFLNAVLQCLSSTRPLRDFCLRRDFRQEVPGGGRAQELTE 260
> Hs6912742
Length=565
Score = 35.0 bits (79), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSE 113
G VGL NL T F ++Q L + PLR++ L D ++ + L+E
Sbjct 210 GHVGLRNLGNTCFLNAVLQCLSSTRPLRDFCLRRDFRQEVPGGGRAQELTE 260
> At2g22310
Length=365
Score = 33.9 bits (76), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 25/100 (25%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query 66 GLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEK-QKYNSVLKTLSELMRKIFNRRNF 124
G N T + ++Q+L P R LL + K ++L L++L +I +++
Sbjct 24 GFENFGNTCYCNSVLQALYFCAPFREQLLEHYANNKADAEENLLTCLADLFSQISSQKKK 83
Query 125 KGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVL 164
G+++P F+Q + ++ + FR D +LLN L
Sbjct 84 TGVIAPKRFVQRLKKQN-ELFRSYMHQDAHEFLNYLLNEL 122
> At4g10570
Length=928
Score = 33.9 bits (76), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 60/250 (24%), Positives = 89/250 (35%), Gaps = 65/250 (26%)
Query 28 TFKYNLNPTYTPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTII 87
T ++L P T ED R S + GK G G GL+NL T F +Q L
Sbjct 264 TSNFSLFPRITSEDDGRDSLSIL-GKGEKG-----GLAGLSNLGNTCFMNSALQCLAHTP 317
Query 88 PLRNYLLLLDLSEKQKYNSVLKTLSELMRKIFNRRNFKG-------------------IV 128
P+ Y L + + + ++ L+ ++ I NF+G V
Sbjct 318 PIVEYFLQDYMLDTYMFYFLVMVLTCILNIILT-LNFQGELAIAFGDLLKKLWSSGRNAV 376
Query 129 SPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD--------------------- 167
+P F + +F Q D L A+LL+ LH D
Sbjct 377 APRAFKTKLA-RFAPQFSGYNQHDSQELLAFLLDGLHEDLNKVKRKPYIELKDSDSRPDD 435
Query 168 -------QKDKKTGGSIIHDCFQGEVLVTDAQSEEGRKT----PFLYLTLDLPPAPIFKD 216
K S+I D QG+ T G+ + PF+YL++ LP
Sbjct 436 EVAEELWNYHKARNDSVIVDVCQGQYKSTLVCPVCGKISITFDPFMYLSVPLP------S 489
Query 217 SLDRNMIPTV 226
+L R+M TV
Sbjct 490 TLTRSMTITV 499
> Hs22065611
Length=1089
Score = 33.5 bits (75), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 51/112 (45%), Gaps = 10/112 (8%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL-LDLSEKQKYNSV------LKTLSELM 115
G GL+NL T F IQ + PL Y + L E + N + K +L+
Sbjct 213 GATGLSNLGNTCFMNSSIQCVSNTQPLTQYFISGRHLYELNRTNPIGMKGHMAKCYGDLV 272
Query 116 RKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
+++++ + V+P + + + + KF +Q D L A+LL+ LH D
Sbjct 273 QELWS--GTQKSVAPLKLRRTI-AKYAPKFDGFQQQDSQELLAFLLDGLHED 321
> HsM7706755
Length=381
Score = 33.5 bits (75), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 0/51 (0%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSE 113
G VGL NL T F ++Q L + PLR++ L D ++ + L+E
Sbjct 26 GHVGLRNLGNTCFLNAVLQCLSSTRPLRDFCLRRDFRQEVPGGGRAQELTE 76
> Hs20483065
Length=749
Score = 33.1 bits (74), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 33/120 (27%), Positives = 52/120 (43%), Gaps = 10/120 (8%)
Query 50 FYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLK 109
FY + + +D G +GL NL T + I+Q+L R+ +L L + Q + L+
Sbjct 159 FYPRLMAKSD--TGKIGLINLGNTCYVNSILQALFMASDFRHCVLRLTENNSQPLMTKLQ 216
Query 110 TLSELMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQK 169
L F + + +SP FL A F G Q D +LL+ LH ++K
Sbjct 217 WLFG-----FLEHSQRPAISPENFLSASWT---PWFSPGTQQDCSEYLKYLLDRLHEEEK 268
> At5g10790
Length=557
Score = 31.6 bits (70), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 37/135 (27%), Positives = 54/135 (40%), Gaps = 16/135 (11%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL----LDLSEKQKYNSVLKTLSELMRKI 118
G GLNNL T F ++Q+L PLRN+ L DL ++ L L + I
Sbjct 175 GLRGLNNLGSTCFMNAVLQALVHAPPLRNFWLSGQHNRDLCPRRTMG--LLCLPCDLDVI 232
Query 119 FNRRNFKGIVSPHE--FLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQ-------K 169
F+ F G +P+ L + EQ D F LL+ +H ++ +
Sbjct 233 FSAM-FSGDRTPYSPAHLLYSWWQHSTNLATYEQQDSHEFFISLLDRIHENEGKSKCLYQ 291
Query 170 DKKTGGSIIHDCFQG 184
D + I H F G
Sbjct 292 DNEECQCITHKAFSG 306
> Hs19923413
Length=1087
Score = 31.2 bits (69), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 27/84 (32%), Positives = 38/84 (45%), Gaps = 12/84 (14%)
Query 38 TPEDVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLL--- 94
TP +V R S + K D A VGL N+ T +F+ +IQSL ++ R +L
Sbjct 147 TPTEVWRDSRNPYDRKRQDKAP-----VGLKNVGNTCWFSAVIQSLFNLLEFRRLVLNYK 201
Query 95 ----LLDLSEKQKYNSVLKTLSEL 114
DL QK + L + EL
Sbjct 202 PPSNAQDLPRNQKEHRNLPFMREL 225
> Hs4758564_2
Length=288
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 51/112 (45%), Gaps = 10/112 (8%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL-LDLSEKQKYNSV------LKTLSELM 115
G GL+NL T F IQ + PL Y + L E + N + K +L+
Sbjct 32 GATGLSNLGNTCFMNSSIQCVSNTQPLTQYFISGRHLYELNRTNPIGMKGHMAKCYGDLV 91
Query 116 RKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
+++++ + V+P + + + + KF +Q D L A+LL+ LH D
Sbjct 92 QELWS--GTQKSVAPLKLRRTIA-KYAPKFDGFQQQDSQELLAFLLDGLHED 140
> Hs4827054
Length=1118
Score = 30.8 bits (68), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 59/139 (42%), Gaps = 7/139 (5%)
Query 35 PTYTPE-DVARLSAEVFYGKSLDGADFLPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYL 93
PT P+ +++RLSA + P GL NL T + I+Q LC L +Y
Sbjct 746 PTCYPKAEISRLSASQIRNLNPVFGGSGPALTGLRNLGNTCYMNSILQCLCNAPHLADYF 805
Query 94 LLLDLSEKQKYNSVLKTLSELMRK--IFNRRNFKG---IVSPHEFLQAVGVESKKKFRIG 148
+ +++L E+ + I + + G +SP +F +G + +F
Sbjct 806 NRNCYQDDINRSNLLGHKGEVAEEFGIIMKALWTGQYRYISPKDFKITIG-KINDQFAGY 864
Query 149 EQSDPLALFAWLLNVLHRD 167
Q D L +L++ LH D
Sbjct 865 SQQDSQELLLFLMDGLHED 883
> Hs14211903
Length=1274
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 29/112 (25%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Query 63 GCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLL-LDLSEKQKYNSV------LKTLSELM 115
G GL+NL T F IQ + PL Y + L E + N + K +L+
Sbjct 402 GATGLSNLGNTCFMNSSIQCVSNTQPLTQYFISGRHLYELNRTNPIGMKGHMAKCYGDLV 461
Query 116 RKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
+++++ + V+P + + + +F +Q D L A+LL+ LH D
Sbjct 462 QELWS--GTQKNVAPLKLRWTIA-KYAPRFNGFQQQDSQELLAFLLDGLHED 510
> At4g15570
Length=525
Score = 30.8 bits (68), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 39/158 (24%), Positives = 71/158 (44%), Gaps = 25/158 (15%)
Query 62 PGCVGLN-NLSKTDFFAVIIQSLCTIIP-LRNYLLLLDLSEKQKYNSVLKTLSELMRKIF 119
PG G NL + +F +I L T+ P L++ L +S YN +KT + +++F
Sbjct 360 PGATGSRVNLDEVEFVLLIYHRLVTMYPELKSSSQLAIIS---PYNYQVKTFKDRFKEMF 416
Query 120 NRRNFKGI-VSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRDQKDKKTGGSII 178
K + ++ + Q VG + K SDPL W + +Q+++
Sbjct 417 GTEAEKVVDINTVDGFQVVGSAATLK------SDPL----WKNLIESAEQRNR------- 459
Query 179 HDCFQGEVLVTDAQSEEGRKTPFLYLTLDLPPAPIFKD 216
F+ + + SEE +T L +++P AP+++D
Sbjct 460 --LFKVSKPLNNFFSEENLETMKLTEDMEIPDAPLYED 495
> CE26210
Length=357
Score = 30.4 bits (67), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 44/118 (37%), Gaps = 17/118 (14%)
Query 61 LPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNSVLKTLSELMRKIFN 120
+PG VGL N+ T F + +Q L L ++ K+ + S + T S L K
Sbjct 20 VPGAVGLFNMGNTCFMSATLQCLFQTPG------LAEVFTKKVFVSKVNTQSRLGSKGVI 73
Query 121 RRNFKGI-----------VSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
F + + P FLQ + G Q D +LL+ LH D
Sbjct 74 SAGFASLSDMIWNGTFTAIRPSRFLQLFSDTVYQPLSDGRQHDASEFQIFLLDALHED 131
> Hs14149817
Length=712
Score = 30.4 bits (67), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 21/39 (53%), Gaps = 0/39 (0%)
Query 62 PGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSE 100
PG GL NL T + ++Q L ++ R L LDL++
Sbjct 270 PGVTGLRNLGNTCYMNSVLQVLSHLLIFRQCFLKLDLNQ 308
> SPCC1494.05c
Length=979
Score = 30.0 bits (66), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 25/114 (21%), Positives = 49/114 (42%), Gaps = 10/114 (8%)
Query 61 LPGCVGLNNLSKTDFFAVIIQSLCTIIPLRNYLLLLDLSEKQKYNS-------VLKTLSE 113
+PG GL+NL T + +Q L LR++ + + ++ V +
Sbjct 307 VPGTCGLSNLGNTCYMNSALQCLTHTRELRDFFTSDEWKNQVNESNPLGMGGQVASIFAS 366
Query 114 LMRKIFNRRNFKGIVSPHEFLQAVGVESKKKFRIGEQSDPLALFAWLLNVLHRD 167
L++ +++ + +P +F +G + G+Q D A+LL+ LH D
Sbjct 367 LIKSLYSPEH--SSFAPRQFKATIGKFNHSFLGYGQQ-DSQEFLAFLLDGLHED 417
Lambda K H
0.321 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5045397172
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40