bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3773_orf1
Length=200
Score E
Sequences producing significant alignments: (Bits) Value
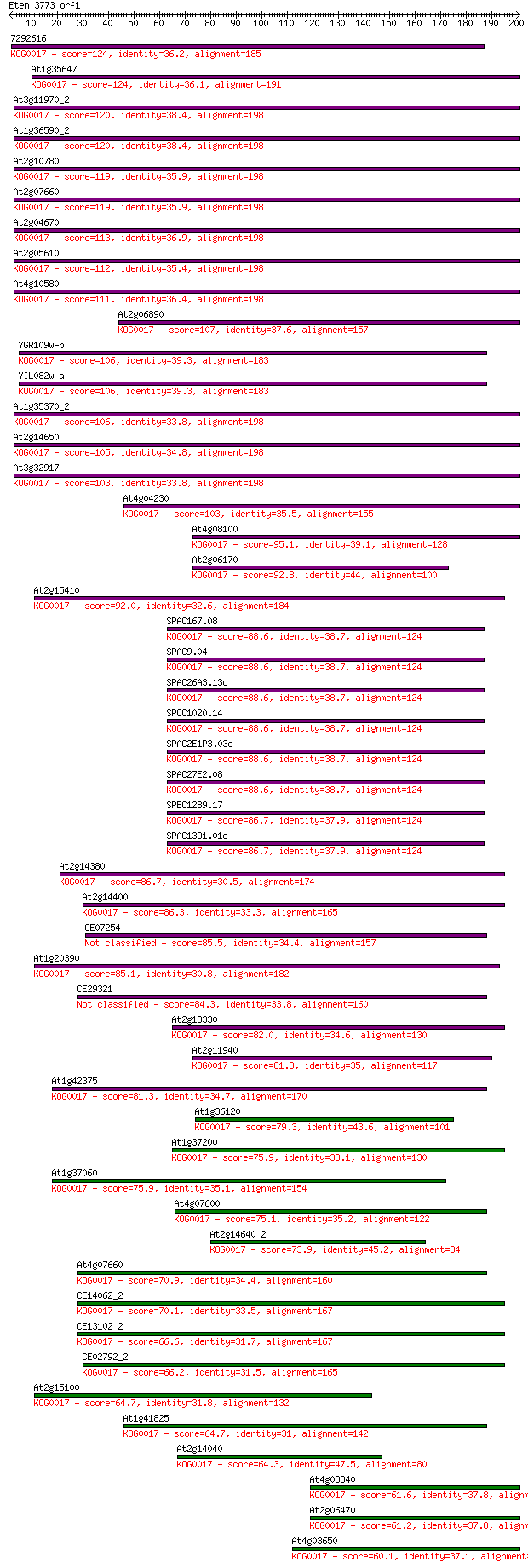
7292616 124 1e-28
At1g35647 124 2e-28
At3g11970_2 120 2e-27
At1g36590_2 120 2e-27
At2g10780 119 4e-27
At2g07660 119 5e-27
At2g04670 113 3e-25
At2g05610 112 6e-25
At4g10580 111 1e-24
At2g06890 107 1e-23
YGR109w-b 106 2e-23
YIL082w-a 106 3e-23
At1g35370_2 106 3e-23
At2g14650 105 9e-23
At3g32917 103 3e-22
At4g04230 103 3e-22
At4g08100 95.1 8e-20
At2g06170 92.8 4e-19
At2g15410 92.0 7e-19
SPAC167.08 88.6 7e-18
SPAC9.04 88.6 7e-18
SPAC26A3.13c 88.6 7e-18
SPCC1020.14 88.6 7e-18
SPAC2E1P3.03c 88.6 7e-18
SPAC27E2.08 88.6 7e-18
SPBC1289.17 86.7 3e-17
SPAC13D1.01c 86.7 3e-17
At2g14380 86.7 3e-17
At2g14400 86.3 4e-17
CE07254 85.5 7e-17
At1g20390 85.1 8e-17
CE29321 84.3 1e-16
At2g13330 82.0 6e-16
At2g11940 81.3 1e-15
At1g42375 81.3 1e-15
At1g36120 79.3 5e-15
At1g37200 75.9 5e-14
At1g37060 75.9 6e-14
At4g07600 75.1 8e-14
At2g14640_2 73.9 2e-13
At4g07660 70.9 2e-12
CE14062_2 70.1 3e-12
CE13102_2 66.6 3e-11
CE02792_2 66.2 4e-11
At2g15100 64.7 1e-10
At1g41825 64.7 1e-10
At2g14040 64.3 2e-10
At4g03840 61.6 9e-10
At2g06470 61.2 1e-09
At4g03650 60.1 3e-09
> 7292616
Length=1062
Score = 124 bits (312), Expect = 1e-28, Method: Composition-based stats.
Identities = 67/189 (35%), Positives = 106/189 (56%), Gaps = 4/189 (2%)
Query 2 HILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGS 61
H + ++P +R +P + T +I KL + I ++S + +V K+ D S
Sbjct 177 HSIKTTDEIPVHKRPFRYSPAEKTEITDQINKLLEQDIIRHSHSPWSAPVFLVPKKLDAS 236
Query 62 GERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDR 117
++K R+V ++R LN T +P+P I IL+ LG A+YFS DL +G+HQI + +DR
Sbjct 237 NKKKWRLVVDFRQLNDKTIKDRYPMPNINEILDKLGRAQYFSALDLASGYHQIEVEPKDR 296
Query 118 WKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCG 177
K AF +V G FE+ MPFGL A ATFQ ++ L G+ + YLDD++++S L
Sbjct 297 SKTAFSAVGGHFEFIRMPFGLSNAPATFQRVMDNVLAEFNGKFCLIYLDDIIVFSTSLQE 356
Query 178 HVSLLRQVL 186
H++ L +
Sbjct 357 HINHLSSIF 365
> At1g35647
Length=1495
Score = 124 bits (310), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 69/195 (35%), Positives = 110/195 (56%), Gaps = 10/195 (5%)
Query 10 LPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSGERKMRMV 69
LP + A YR P + ++++ +L + G I + S C+ +++ + DGS RM
Sbjct 518 LPNRPA-YRTNPVETKELEKQVTELMERGHIRESMSP-CAVPVLLVPKKDGS----WRMC 571
Query 70 ANYRALNSLTF----PLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSV 125
+ RA+N++T P+P + +L+ L G+ FS DL +G+HQI M D WK AF ++
Sbjct 572 VDCRAINNITVKYRHPIPRLDDMLDELHGSSIFSKVDLKSGYHQIRMKEGDEWKTAFKTI 631
Query 126 LGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQV 185
G +E+ VMPFGL A +TF +N L+ +G+ VI Y DD+L+YS L HV L+ V
Sbjct 632 QGLYEWLVMPFGLTNAPSTFMRLMNHVLRAFIGRFVIVYFDDILVYSKSLEEHVEHLKMV 691
Query 186 LSIFLRYQFYPKFRK 200
L + + + Y +K
Sbjct 692 LEVLRKEKLYANLKK 706
> At3g11970_2
Length=958
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 76/206 (36%), Positives = 108/206 (52%), Gaps = 17/206 (8%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSD----NGWIGLTYSSICSRTIMVEKRD 58
I L+ G P YR + Q K EI KL + NG + + S S ++V+K+D
Sbjct 52 IKLLEGSNPVNQRPYRYSIHQ----KNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKD 107
Query 59 DGSGERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAA 114
R+ +YR LN +T FP+P I+ +++ LGGA FS DL AG+HQ+ M
Sbjct 108 GT-----WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDP 162
Query 115 EDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPD 174
+D K AF + G FEY VMPFGL A ATFQ +N + L + V+ + DD+L+YS
Sbjct 163 DDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSS 222
Query 175 LCGHVSLLRQVLSIFLRYQFYPKFRK 200
L H L+QV + + + K K
Sbjct 223 LEEHRQHLKQVFEVMRANKLFAKLSK 248
> At1g36590_2
Length=958
Score = 120 bits (301), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 76/206 (36%), Positives = 108/206 (52%), Gaps = 17/206 (8%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSD----NGWIGLTYSSICSRTIMVEKRD 58
I L+ G P YR + Q K EI KL + NG + + S S ++V+K+D
Sbjct 52 IKLLEGSNPVNQRPYRYSIHQ----KNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKD 107
Query 59 DGSGERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAA 114
R+ +YR LN +T FP+P I+ +++ LGGA FS DL AG+HQ+ M
Sbjct 108 GT-----WRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQVRMDP 162
Query 115 EDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPD 174
+D K AF + G FEY VMPFGL A ATFQ +N + L + V+ + DD+L+YS
Sbjct 163 DDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILVYSSS 222
Query 175 LCGHVSLLRQVLSIFLRYQFYPKFRK 200
L H L+QV + + + K K
Sbjct 223 LEEHRQHLKQVFEVMRANKLFAKLSK 248
> At2g10780
Length=1611
Score = 119 bits (298), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 71/202 (35%), Positives = 107/202 (52%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G P A YR P + K+++ +L D G+I + S + + V+K+D GS
Sbjct 651 IELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKD-GS- 708
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +YR LN +T +PLP I +++ LGGA++FS DL +G+HQI + D
Sbjct 709 ---FRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVR 765
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + FE+ VMPFGL A A F +N + L + VI +++D+L+YS H
Sbjct 766 KTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAH 825
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR VL ++ + K K
Sbjct 826 QEHLRAVLERLREHELFAKLSK 847
> At2g07660
Length=949
Score = 119 bits (297), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 71/202 (35%), Positives = 107/202 (52%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G P A YR P + K+++ +L G+I + S + + V+K+D GS
Sbjct 146 IELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKD-GS- 203
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +YR LN +T +PLP I +++ LGGA++FS DL +G+HQI + D
Sbjct 204 ---FRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVR 260
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPFGL A A F +N + L + VI ++DD+L++S H
Sbjct 261 KTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAH 320
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR VL ++ + K K
Sbjct 321 QEHLRAVLERLREHELFAKLSK 342
> At2g04670
Length=1411
Score = 113 bits (282), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 73/202 (36%), Positives = 105/202 (51%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G P A YR P + T K+++ L G+I + S + + V+K+D GS
Sbjct 509 IELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKD-GS- 566
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +YR LN +T +PLP I +L+ L GA FS DL +G+HQI +A D
Sbjct 567 ---FRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVR 623
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPF L A A F +N+ Q L + VI ++DD+L+YS H
Sbjct 624 KTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEH 683
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR+V+ + + K K
Sbjct 684 EVHLRRVMEKLREQKLFAKLSK 705
> At2g05610
Length=780
Score = 112 bits (279), Expect = 6e-25, Method: Compositional matrix adjust.
Identities = 70/206 (33%), Positives = 106/206 (51%), Gaps = 17/206 (8%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDN----GWIGLTYSSICSRTIMVEKRD 58
I L+ P YR Q K EI K+ D+ G I + S S ++V+K+D
Sbjct 35 IELLEDSNPVNQRPYRYVVHQ----KNEIDKIVDDMLASGTIQASSSPYASPVVLVKKKD 90
Query 59 DGSGERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAA 114
R+ +YR LN +T FP+P I+ +++ LGG+ +S DL AG+HQ+ M
Sbjct 91 G-----TWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYSKIDLRAGYHQVRMDP 145
Query 115 EDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPD 174
D K AF + G +EY VMPFGL A A+FQ+ +N++ + L + V+ + DD+LIYS
Sbjct 146 LDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRKFVLVFFDDILIYSTS 205
Query 175 LCGHVSLLRQVLSIFLRYQFYPKFRK 200
+ H L V + + + K K
Sbjct 206 MEEHKKHLEAVFEVMRVHHLFAKMSK 231
> At4g10580
Length=1240
Score = 111 bits (277), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 72/202 (35%), Positives = 104/202 (51%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G P A YR P + K+++ L G+I + S + + V+K+D GS
Sbjct 483 IELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKD-GS- 540
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +YR LN +T +PLP I +L+ L GA FS DL +G+HQI +A D
Sbjct 541 ---FRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVR 597
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPFGL A A F +N+ Q L + VI ++DD+L+YS
Sbjct 598 KTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQ 657
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR+V+ + + K K
Sbjct 658 EVHLRRVMEKLREQKLFAKLSK 679
> At2g06890
Length=1215
Score = 107 bits (268), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 59/161 (36%), Positives = 90/161 (55%), Gaps = 8/161 (4%)
Query 44 YSSICSRTIMVEKRDDGSGERKMRMVANYRALNSLTF----PLPPIQTILEMLGGAKYFS 99
Y + T ++++ DGS RM + RA+N++T P+P + +L+ L G+ FS
Sbjct 451 YRTNPVETKELQRQKDGS----WRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFS 506
Query 100 TFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQ 159
DL +G+HQI M D WK AF + G +E+ VMPFGL A +TF +N L+ +G
Sbjct 507 KIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGI 566
Query 160 GVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQFYPKFRK 200
VI Y DD+L+YS L H+ L VL++ + + Y +K
Sbjct 567 FVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKK 607
> YGR109w-b
Length=1547
Score = 106 bits (265), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 72/210 (34%), Positives = 104/210 (49%), Gaps = 34/210 (16%)
Query 5 LVRGKLPAKSAIYRKTPDQHTF-------------------HKQEI----AKLSDNGWIG 41
++R LP + A P +H ++QEI KL DN +I
Sbjct 567 IIRNDLPPRPADINNIPVKHDIEIKPGARLPRLQPYHVTEKNEQEINKIVQKLLDNKFI- 625
Query 42 LTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKY 97
+ S CS +++ + DG+ R+ +YR LN T FPLP I +L +G A+
Sbjct 626 VPSKSPCSSPVVLVPKKDGT----FRLCVDYRTLNKATISDPFPLPRIDNLLSRIGNAQI 681
Query 98 FSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLL 157
F+T DL +G+HQI M +DR+K AF++ G +EY VMPFGL A +TF + + L
Sbjct 682 FTTLDLHSGYHQIPMEPKDRYKTAFVTPSGKYEYTVMPFGLVNAPSTFARYMADTFRDL- 740
Query 158 GQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
+ V YLDD+LI+S H L VL
Sbjct 741 -RFVNVYLDDILIFSESPEEHWKHLDTVLE 769
> YIL082w-a
Length=1498
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 72/210 (34%), Positives = 104/210 (49%), Gaps = 34/210 (16%)
Query 5 LVRGKLPAKSAIYRKTPDQHTF-------------------HKQEI----AKLSDNGWIG 41
++R LP + A P +H ++QEI KL DN +I
Sbjct 593 IIRNDLPPRPADINNIPVKHDIEIKPGARLPRLQPYHVTEKNEQEINKIVQKLLDNKFI- 651
Query 42 LTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKY 97
+ S CS +++ + DG+ R+ +YR LN T FPLP I +L +G A+
Sbjct 652 VPSKSPCSSPVVLVPKKDGT----FRLCVDYRTLNKATISDPFPLPRIDNLLSRIGNAQI 707
Query 98 FSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLL 157
F+T DL +G+HQI M +DR+K AF++ G +EY VMPFGL A +TF + + L
Sbjct 708 FTTLDLHSGYHQIPMEPKDRYKTAFVTPSGKYEYTVMPFGLVNAPSTFARYMADTFRDL- 766
Query 158 GQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
+ V YLDD+LI+S H L VL
Sbjct 767 -RFVNVYLDDILIFSESPEEHWKHLDTVLE 795
> At1g35370_2
Length=923
Score = 106 bits (265), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 67/202 (33%), Positives = 104/202 (51%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L+ G P YR Q + + + +G I ++ S S ++V+K+D
Sbjct 48 IKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKDG--- 104
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +Y LN +T F +P I+ +++ LGG+ FS DL AG+HQ+ M +D
Sbjct 105 --TWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQ 162
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FEY VM FGL A ATFQ+ +N+ + L + V+ + DD+LIYS + H
Sbjct 163 KTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEH 222
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR V + ++ + K K
Sbjct 223 KEHLRLVFEVMRLHKLFAKGSK 244
> At2g14650
Length=1328
Score = 105 bits (261), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 69/202 (34%), Positives = 99/202 (49%), Gaps = 21/202 (10%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G P A YR P + K+++ L ++ K+ DGS
Sbjct 470 IELEPGTAPLSKAPYRMAPAEMAELKKQLEDL-------------LGAPVLFVKKKDGS- 515
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
R+ +YR LN +T +PLP I +L+ L GA FS DL +G+H I +A D
Sbjct 516 ---FRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVR 572
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPFGL A A F +N+ Q +L + VI ++DD+L+YS L H
Sbjct 573 KTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEH 632
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR+V+ + + K K
Sbjct 633 EVHLRRVMEKLREQKLFAKLSK 654
> At3g32917
Length=757
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 67/202 (33%), Positives = 102/202 (50%), Gaps = 9/202 (4%)
Query 3 ILLVRGKLPAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSG 62
I L G A YR P + K+++ L G+I + +S+ ++ K+ DGS
Sbjct 546 IELEPGTASLSKAPYRMAPAEMAELKKQLEDLLGKGFIRPS-TSLWGAPVLFVKKKDGS- 603
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
+ +YR LN +T +PLP I +L+ L G FS DL +G+HQI +A D
Sbjct 604 ---FHLCIDYRGLNRVTVKNKYPLPRIDELLDQLRGGTCFSKIDLTSGYHQIPIAEGDVR 660
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPFGL A A+F +N+ L + VI ++DD+L+YS H
Sbjct 661 KTAFRTRYGIFEFVVMPFGLTNAPASFMRLMNSVFYEFLDEFVIIFIDDILVYSKSPEEH 720
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
+R+V+ + + K K
Sbjct 721 EVRMRRVIEKLRELKLFAKLSK 742
> At4g04230
Length=315
Score = 103 bits (256), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 55/159 (34%), Positives = 86/159 (54%), Gaps = 8/159 (5%)
Query 46 SICSRTIMVEKRDDGSGERKMRMVANYRALNSLTF----PLPPIQTILEMLGGAKYFSTF 101
S C+ +++ + DG+ RM + RA+N++T P+P + +L+ L GA FS
Sbjct 135 SPCAVPVLLVPKKDGT----WRMCLDCRAINNITIKYRHPIPRLYDMLDELSGAIIFSKV 190
Query 102 DLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGV 161
DL +G+HQ+ M D WK AF + G +E VMPFGL A +TF +N L+ +G+ V
Sbjct 191 DLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRSFIGKFV 250
Query 162 IAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQFYPKFRK 200
+ Y DD+LIY+ H+ L +L + Y +K
Sbjct 251 VVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKK 289
> At4g08100
Length=1054
Score = 95.1 bits (235), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 50/132 (37%), Positives = 73/132 (55%), Gaps = 4/132 (3%)
Query 73 RALNSLTF----PLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGP 128
RA+N++T P+P + L+ L G+ FS DL +G+HQ M D WK A +
Sbjct 529 RAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQRL 588
Query 129 FEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSI 188
+E+ VMPFGL A TF +N L+ +G VI Y DD+L+YS +L HV L+ VL +
Sbjct 589 YEWLVMPFGLTNAPNTFMRLMNHVLRKHIGVFVIVYFDDILVYSKNLEYHVMHLKLVLDL 648
Query 189 FLRYQFYPKFRK 200
+ + Y +K
Sbjct 649 LRKEKLYANLKK 660
> At2g06170
Length=587
Score = 92.8 bits (229), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 44/104 (42%), Positives = 64/104 (61%), Gaps = 4/104 (3%)
Query 73 RALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGP 128
+A+N +T FP+P + +L+ L G+K FS DL +G+HQI + D WK F S G
Sbjct 401 KAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRPGDEWKTDFKSKDGL 460
Query 129 FEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYS 172
+E++VMPFG+ A +TF +N L+ G V+ Y DD+LIYS
Sbjct 461 YEWQVMPFGMSNAPSTFMRLMNQILRPFTGSFVVVYFDDILIYS 504
> At2g15410
Length=1787
Score = 92.0 bits (227), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 60/188 (31%), Positives = 90/188 (47%), Gaps = 8/188 (4%)
Query 11 PAKSAIYRKTPDQHTFHKQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSGERKMRMVA 70
P K + PD+ +E+ KL D G I R +V K+ +G K R+
Sbjct 782 PLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVEVRYPDWLRNPVVVKKKNG----KWRVCI 837
Query 71 NYRALNSL----TFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVL 126
++ LN +FPLP I ++E G + S D +G++QI M DR K F++
Sbjct 838 DFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMDAFSGYNQILMHQNDREKTVFITDQ 897
Query 127 GPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVL 186
G + Y+VMPFGLK A AT+ +N L + Y+DD+L+ S H++ LRQ
Sbjct 898 GTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSMEVYIDDMLVKSLRAEEHITHLRQCF 957
Query 187 SIFLRYQF 194
+ RY
Sbjct 958 QVLNRYNM 965
> SPAC167.08
Length=1214
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 340 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 399
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 400 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 459
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 460 VKHVKDVL 467
> SPAC9.04
Length=1333
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPAC26A3.13c
Length=1333
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPCC1020.14
Length=1333
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPAC2E1P3.03c
Length=1333
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPAC27E2.08
Length=1333
Score = 88.6 bits (218), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 48/128 (37%), Positives = 69/128 (53%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L V+ Y+DD+LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISTAPAHFQYFINTILGEAKESHVVCYMDDILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPBC1289.17
Length=1333
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 70/128 (54%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L + V+ Y+D++LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISIAPAHFQYFINTILGEVKESHVVCYMDNILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> SPAC13D1.01c
Length=1333
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 47/128 (36%), Positives = 70/128 (54%), Gaps = 4/128 (3%)
Query 63 ERKMRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRW 118
E +RMV +Y+ LN +PLP I+ +L + G+ F+ DL + +H I + D
Sbjct 459 EGTLRMVVDYKPLNKYVKPNIYPLPLIEQLLAKIQGSTIFTKLDLKSAYHLIRVRKGDEH 518
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K+AF G FEY VMP+G+ A A FQ IN L + V+ Y+D++LI+S H
Sbjct 519 KLAFRCPRGVFEYLVMPYGISIAPAHFQYFINTILGEVKESHVVCYMDNILIHSKSESEH 578
Query 179 VSLLRQVL 186
V ++ VL
Sbjct 579 VKHVKDVL 586
> At2g14380
Length=764
Score = 86.7 bits (213), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 53/179 (29%), Positives = 89/179 (49%), Gaps = 10/179 (5%)
Query 21 PDQHTFHKQEIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSL- 78
P++ E+ KL D G I + Y + ++V+K++D K R+ ++ LN
Sbjct 507 PERSKAVNDEVDKLLDAGSIVEVKYPEWLANPVVVKKKND-----KWRVCIDFTDLNKAC 561
Query 79 ---TFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMP 135
+FPLP I ++E G + S D +G++QI M +D+ K +F+ G + Y+VMP
Sbjct 562 PKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQIPMHKDDQEKTSFIIDRGTYCYKVMP 621
Query 136 FGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQF 194
FGLK A +Q +N LG+ + Y+DD+L+ S H+ L+ +Y
Sbjct 622 FGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDMLVKSTRSADHIDHLKACFETLNKYNM 680
> At2g14400
Length=1466
Score = 86.3 bits (212), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 55/170 (32%), Positives = 89/170 (52%), Gaps = 10/170 (5%)
Query 30 EIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSL----TFPLPP 84
E+ KL G I + Y + T++V+K++ K R+ ++ LN +FPLP
Sbjct 483 EVDKLLKIGSIREVQYPDWLANTVVVKKKNG-----KDRVCIDFTDLNKACPKDSFPLPH 537
Query 85 IQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALAT 144
I ++E G + S D +G++QI M ED+ K F++ G + Y+VMPFGL+ A AT
Sbjct 538 IDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGAT 597
Query 145 FQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQF 194
+ +N +G+ + Y+DD+LI S HV L + +I +YQ
Sbjct 598 YPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQM 647
> CE07254
Length=2175
Score = 85.5 bits (210), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 54/161 (33%), Positives = 86/161 (53%), Gaps = 9/161 (5%)
Query 31 IAKLSDNGWIGLTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSLT----FPLPPIQ 86
I K+ + I + S S ++V+K+D GS +RM +YR +N + PLP I+
Sbjct 951 IQKMLNQKVIRESKSPWSSPVVLVKKKD-GS----IRMCIDYRKVNKVVKNNAHPLPNIE 1005
Query 87 TILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQ 146
L+ L G K ++ FD+ AGF QI + + + AF FE+ V+PFGL + A FQ
Sbjct 1006 ATLQSLAGKKLYTVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGLVISPALFQ 1065
Query 147 ANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
+ + LLG Y+DD+LI S D+ H+ +++ L+
Sbjct 1066 GTMEEIIGDLLGVCAFVYVDDLLIASKDMEQHLQDVKEALT 1106
> At1g20390
Length=1791
Score = 85.1 bits (209), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 56/187 (29%), Positives = 93/187 (49%), Gaps = 10/187 (5%)
Query 11 PAKSAIYRKTPDQHTFHKQEIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMV 69
P K + P++ +E+ KL G I + Y + ++V+K++ K R+
Sbjct 777 PVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNG-----KWRVC 831
Query 70 ANYRALNSL----TFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSV 125
+Y LN ++PLP I ++E G S D +G++QI M +D+ K +F++
Sbjct 832 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 891
Query 126 LGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQV 185
G + Y+VM FGLK A AT+Q +N L +G+ V Y+DD+L+ S HV L +
Sbjct 892 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 951
Query 186 LSIFLRY 192
+ Y
Sbjct 952 FDVLNTY 958
> CE29321
Length=2186
Score = 84.3 bits (207), Expect = 1e-16, Method: Composition-based stats.
Identities = 54/164 (32%), Positives = 88/164 (53%), Gaps = 9/164 (5%)
Query 28 KQEIAKLSDNGWIGLTYSSICSRTIMVEKRDDGSGERKMRMVANYRALNSLT----FPLP 83
++ I K+ + I + S S ++V+K+D GS +RM +YR +N + PLP
Sbjct 959 RKMIQKMLNQKVIRESKSPWSSPVVLVKKKD-GS----IRMCIDYRKVNKVVKNNAHPLP 1013
Query 84 PIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALA 143
I+ L+ L G K ++ FD+ AGF QI + + + AF FE+ V+PFGL + A
Sbjct 1014 NIEATLQSLAGKKLYTVFDMIAGFWQIPLDEKSKEITAFAIGSELFEWNVLPFGLVISPA 1073
Query 144 TFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
FQ + + LLG Y+DD+LI S D+ H+ +++ L+
Sbjct 1074 LFQGTMEEIIGDLLGVCAFVYVDDLLIASKDMEQHLQDVKEALT 1117
> At2g13330
Length=889
Score = 82.0 bits (201), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 45/134 (33%), Positives = 72/134 (53%), Gaps = 4/134 (2%)
Query 65 KMRMVANYRALNSL----TFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKM 120
K R+ ++R LN +FPLP I ++E + S D G++QI M + + K
Sbjct 293 KWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDGQEKT 352
Query 121 AFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVS 180
AF++ G + Y+VMPFGLK A T+Q +N LG+ + Y+DD+L+ S HV
Sbjct 353 AFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEKDHVP 412
Query 181 LLRQVLSIFLRYQF 194
LR+ I ++++
Sbjct 413 QLRECFKILIKFEM 426
> At2g11940
Length=1212
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/117 (35%), Positives = 65/117 (55%), Gaps = 0/117 (0%)
Query 73 RALNSLTFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYR 132
R L + FPLP I +E G + S D +G++QI M +D+ K +F++ G + Y+
Sbjct 424 RLLKADCFPLPHIDRPIESTTGQEMLSFMDAFSGYNQIMMNPDDQEKTSFITERGTYYYK 483
Query 133 VMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIF 189
VMPFGLK AT+Q +N + LLG+ + Y+DD+L+ + H+ +Q I
Sbjct 484 VMPFGLKNVGATYQLLVNIMFKDLLGKTMEVYIDDMLVKTAFSASHLDHFQQCFDIL 540
> At1g42375
Length=1773
Score = 81.3 bits (199), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 59/188 (31%), Positives = 88/188 (46%), Gaps = 18/188 (9%)
Query 18 RKTPDQHTFHKQEIAKL---------SDNGWIGLTYSSICSRTIMVEK--RDDGSGERKM 66
R P+ K+EI KL SD+ W+ + ++V K +D+ R +
Sbjct 875 RLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKDGMIVVKNEKDELIPTRTI 934
Query 67 ---RMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWK 119
RM +YR LN+ + FPLP I +LE L Y+ D +GF QI + D+ K
Sbjct 935 TGHRMCIDYRKLNAASRKDHFPLPFIDQMLERLANHPYYCFLDGYSGFFQIPIHPNDQEK 994
Query 120 MAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHV 179
F G F Y+ MPFGL A ATFQ + + L+ + V ++DD +Y P +
Sbjct 995 TTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIKEMVEVFMDDFSVYGPSFSSCL 1054
Query 180 SLLRQVLS 187
L +VL+
Sbjct 1055 LNLGRVLT 1062
> At1g36120
Length=1235
Score = 79.3 bits (194), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 44/107 (41%), Positives = 60/107 (56%), Gaps = 6/107 (5%)
Query 74 ALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPF 129
LN +T +PLP I +L+ L GA FS DL G+HQ +A D K AF + G F
Sbjct 575 GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAFRTRYGHF 634
Query 130 EYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIY--SPD 174
E+ VMPFGL A +N+ Q L + VI ++DD+L+Y SP+
Sbjct 635 EFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPE 681
> At1g37200
Length=1564
Score = 75.9 bits (185), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 43/134 (32%), Positives = 71/134 (52%), Gaps = 8/134 (5%)
Query 65 KMRMVANYRALNSL----TFPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKM 120
K R+ ++ LN +FPLP I +++ G + S D +G++QI M +D+ K
Sbjct 735 KWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEKT 794
Query 121 AFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVS 180
AF++ Y+VMPFGLK AT+Q +N LG+ + Y+DD+L+ S H+
Sbjct 795 AFITDC----YKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHLP 850
Query 181 LLRQVLSIFLRYQF 194
LR+ I +++
Sbjct 851 QLRECFKILNKFEM 864
> At1g37060
Length=1734
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 54/172 (31%), Positives = 78/172 (45%), Gaps = 18/172 (10%)
Query 18 RKTPDQHTFHKQEIAKL---------SDNGWIGLTYS--SICSRTIMVEKRDDGSGERKM 66
R P+ K+EI KL SD+ W+ + T++ +D+ R +
Sbjct 906 RLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTI 965
Query 67 ---RMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWK 119
RM YR LN + FPLP I +LE L Y+ D +GF QI + D+ K
Sbjct 966 TGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGK 1025
Query 120 MAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIY 171
F G F Y+ MPFGL A ATFQ + + L+ + V ++DD +Y
Sbjct 1026 TTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVY 1077
> At4g07600
Length=630
Score = 75.1 bits (183), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 43/126 (34%), Positives = 62/126 (49%), Gaps = 4/126 (3%)
Query 66 MRMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMA 121
+RM +YR LN+ + FPLP +LE L Y+ D +GF QI + D K
Sbjct 355 LRMCIDYRNLNAASRNDHFPLPFTDQMLERLANHPYYCFLDGYSGFFQIPIHPNDHEKTT 414
Query 122 FLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSL 181
F G F Y MPFGL A ATFQ + + ++ + V ++DD +Y P +
Sbjct 415 FTCPYGTFAYERMPFGLCNAPATFQRCMTSIFSDIIEEMVEVFMDDFSVYGPSFSSCLLN 474
Query 182 LRQVLS 187
L +VL+
Sbjct 475 LGRVLT 480
> At2g14640_2
Length=492
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 52/84 (61%), Gaps = 0/84 (0%)
Query 80 FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLK 139
FP+P + +L+ L GA YF+ DL AG+ Q+ M + D K+AF + G +EY VMPFGL
Sbjct 100 FPIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHSPDIPKIAFQTHNGHYEYLVMPFGLC 159
Query 140 GALATFQANINAYLQHLLGQGVIA 163
A +TFQA +N LL + V A
Sbjct 160 NAPSTFQALMNEIFWPLLSRSVAA 183
> At4g07660
Length=724
Score = 70.9 bits (172), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 55/178 (30%), Positives = 81/178 (45%), Gaps = 18/178 (10%)
Query 28 KQEIAKLSDNGWIGLTYSSI------C-----SRTIMVEKRDDGSGERKM---RMVANYR 73
K+EI KL D G I S C T++ ++D+ R + R+ +YR
Sbjct 366 KKEILKLLDAGVIYPISDSTWVFPVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCIDYR 425
Query 74 ALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPF 129
LN+ + FPLP +LE L Y D +GF QI + D+ K F G F
Sbjct 426 KLNAASRKDHFPLPFTNQMLEGLANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPYGTF 485
Query 130 EYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
Y+ MPFGL A TFQ + + L+ + V ++DD +Y P + L +VL+
Sbjct 486 AYKRMPFGLCNAPTTFQRCMTSIFSDLIEKMVEVFMDDFSVYGPSFSSCLLNLGRVLT 543
> CE14062_2
Length=812
Score = 70.1 bits (170), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 56/174 (32%), Positives = 94/174 (54%), Gaps = 14/174 (8%)
Query 28 KQEIAKLSDNGWIG-LTYSSICSRTIMVEKRDDGSGERKMRMVANYRAL---NSLTFPLP 83
+QE+ +L D + +T+S + +++ K+D G K+R+ A+++ NSL +
Sbjct 58 EQELNRLLDLEVLEPITHSDWAAPIVVIRKKDTG----KVRVCADFKCSGLNNSLIEEIH 113
Query 84 PIQTILEMLGGAK--YFSTFDLGAGFHQISMAAEDRWKMAFLSV-LGPFEYRVMPFGLKG 140
P+ T ++ G + FS DL + QI++ +E + K+A ++ G F+YR M FGLK
Sbjct 114 PLPTSDDLFGTLQGCIFSKIDLKDAYLQIALDSESQ-KLAVINTHKGLFKYRRMTFGLKP 172
Query 141 ALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQF 194
A A FQ I+ + L GV AYLDDV++ + L H +L ++L Y F
Sbjct 173 APAKFQKIIDKMIAGL--PGVAAYLDDVIVSANSLEEHEKVLHELLKRIKDYGF 224
> CE13102_2
Length=813
Score = 66.6 bits (161), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 53/175 (30%), Positives = 92/175 (52%), Gaps = 16/175 (9%)
Query 28 KQEIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMVANYR------ALNSLTF 80
++E+ +L + G I +TY+ + ++++K+ G+G K+R+ A+++ AL
Sbjct 58 EKELDRLQEMGVIVPITYAKWAAPIVVIKKK--GTG--KIRVCADFKCSGLNAALKDEFH 113
Query 81 PLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSV-LGPFEYRVMPFGLK 139
PLP + I L G Y S DL + Q+ + E + K+A ++ G F+Y M FGLK
Sbjct 114 PLPTSEDIFSRLKGTVY-SQIDLKDAYLQVELDEEAQ-KLAVINTHRGIFKYLRMTFGLK 171
Query 140 GALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQF 194
A A+FQ ++ + L GV YLDD++I + + H +LR++ Y F
Sbjct 172 PAPASFQKIMDKMVSGLT--GVAVYLDDIIISASSIEEHEKILRELFERIKEYGF 224
> CE02792_2
Length=634
Score = 66.2 bits (160), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 52/173 (30%), Positives = 88/173 (50%), Gaps = 16/173 (9%)
Query 30 EIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMVANYR------ALNSLTFPL 82
E+ +L + G I +TY+ + ++++K+ G K+R+ A+++ AL PL
Sbjct 60 ELNRLQEMGVIVPITYAKWAAPIVVIKKKGTG----KIRVCADFKCSGLNAALKDEFHPL 115
Query 83 PPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSV-LGPFEYRVMPFGLKGA 141
P + I L G Y S DL + Q+ + E + K+A ++ G F+Y M FGLK A
Sbjct 116 PTSEDIFSRLKGTVY-SQIDLKDAYLQVELDEEAQ-KLAVINTHRGIFKYLRMTFGLKPA 173
Query 142 LATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLSIFLRYQF 194
A+FQ ++ + L GV Y DD++I + + H +LR++ F Y F
Sbjct 174 PASFQKIMDKMVSGLT--GVAVYWDDIIISASSIEEHEKILRELFERFKEYGF 224
> At2g15100
Length=1329
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/137 (30%), Positives = 70/137 (51%), Gaps = 10/137 (7%)
Query 11 PAKSAIYRKTPDQHTFHKQEIAKLSDNGWI-GLTYSSICSRTIMVEKRDDGSGERKMRMV 69
P K + PD+ E+ +L + G I + Y + ++V+K++ K R+
Sbjct 432 PVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLANPVVVKKKNG-----KWRVC 486
Query 70 ANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSV 125
++ LN FPLP I ++E G + S D +G++QI M ED+ K +F++
Sbjct 487 VDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQILMNPEDQEKTSFITE 546
Query 126 LGPFEYRVMPFGLKGAL 142
G + Y+VMPFGLK A+
Sbjct 547 CGTYYYKVMPFGLKNAV 563
> At1g41825
Length=884
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 44/155 (28%), Positives = 69/155 (44%), Gaps = 13/155 (8%)
Query 46 SICSRTIMVEKRDDGSGERKMRMVAN---------YRALNSLT----FPLPPIQTILEML 92
+C I ++ S E + R+ N + LN+ T FPL I +LE L
Sbjct 479 DLCMHRIHLKDESKPSVEHRRRLNPNLKDAVKKEIMKKLNAATRKNHFPLQFIDQLLERL 538
Query 93 GGAKYFSTFDLGAGFHQISMAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAY 152
KY+ D +GF QI + +D+ K F G F Y MPFGL A A F+ + +
Sbjct 539 SNHKYYCVLDGYSGFFQIPIHPDDQEKTMFTCPYGTFAYSRMPFGLCNAPAIFERCMMSI 598
Query 153 LQHLLGQGVIAYLDDVLIYSPDLCGHVSLLRQVLS 187
++ + ++DD +Y + LR+VL+
Sbjct 599 FTDMIENFIEVFMDDFSVYGSSFEACLENLRKVLA 633
> At2g14040
Length=1048
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 44/84 (52%), Gaps = 4/84 (4%)
Query 67 RMVANYRALNSLT----FPLPPIQTILEMLGGAKYFSTFDLGAGFHQISMAAEDRWKMAF 122
RM +YR LNS T FPL I +LE L Y+ D GF QI + +D+ K F
Sbjct 542 RMCIDYRKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTF 601
Query 123 LSVLGPFEYRVMPFGLKGALATFQ 146
G F YR MPFGL A ATFQ
Sbjct 602 TCPYGTFAYRRMPFGLCNAPATFQ 625
> At4g03840
Length=973
Score = 61.6 bits (148), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 43/82 (52%), Gaps = 0/82 (0%)
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AFL+ G FE+ VMPFGL A F +N + L + VI ++DD+L+YS H
Sbjct 290 KTAFLTRYGHFEFVVMPFGLTNAPTAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSWEAH 349
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR VL ++ + K K
Sbjct 350 QEHLRAVLEQLREHELFAKLSK 371
> At2g06470
Length=899
Score = 61.2 bits (147), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/82 (37%), Positives = 43/82 (52%), Gaps = 0/82 (0%)
Query 119 KMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIYSPDLCGH 178
K AF + G FE+ VMPFGL A A F +N + L + VI ++DD+L+YS H
Sbjct 290 KTAFRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVYSKSWEAH 349
Query 179 VSLLRQVLSIFLRYQFYPKFRK 200
LR VL ++ + K K
Sbjct 350 QEHLRAVLERLREHELFAKLSK 371
> At4g03650
Length=839
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 47/89 (52%), Gaps = 0/89 (0%)
Query 112 MAAEDRWKMAFLSVLGPFEYRVMPFGLKGALATFQANINAYLQHLLGQGVIAYLDDVLIY 171
+A D K AF + G FE+ VMPFGL A A F +N+ Q L + VI ++DD+L+Y
Sbjct 516 LAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVY 575
Query 172 SPDLCGHVSLLRQVLSIFLRYQFYPKFRK 200
S H LR+V+ + + K K
Sbjct 576 SKSPEEHEVHLRRVMEKLREQKLFAKLSK 604
Lambda K H
0.326 0.140 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3479363422
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40