bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3783_orf2
Length=209
Score E
Sequences producing significant alignments: (Bits) Value
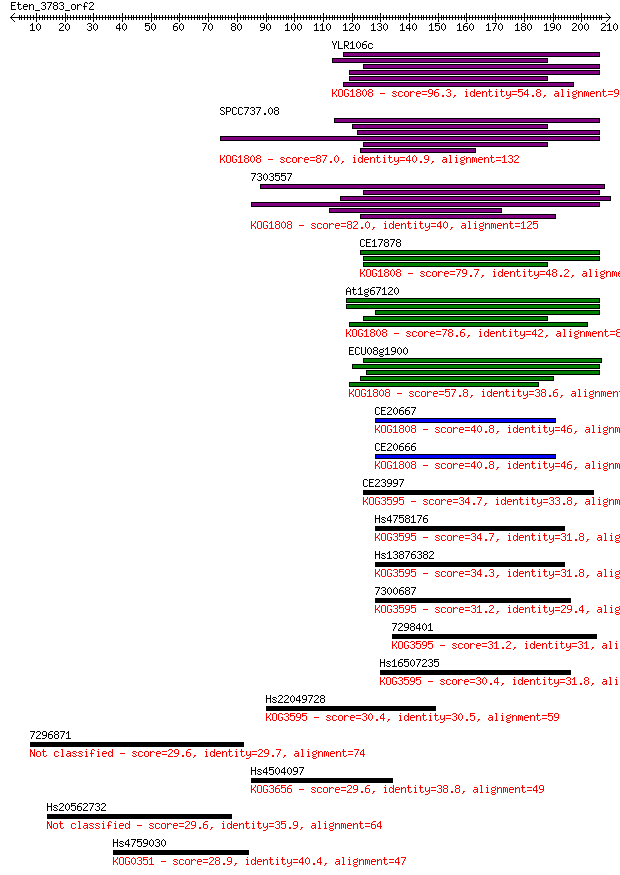
YLR106c 96.3 4e-20
SPCC737.08 87.0 3e-17
7303557 82.0 9e-16
CE17878 79.7 4e-15
At1g67120 78.6 9e-15
ECU08g1900 57.8 2e-08
CE20667 40.8 0.002
CE20666 40.8 0.002
CE23997 34.7 0.13
Hs4758176 34.7 0.16
Hs13876382 34.3 0.19
7300687 31.2 1.5
7298401 31.2 1.7
Hs16507235 30.4 2.5
Hs22049728 30.4 2.6
7296871 29.6 4.5
Hs4504097 29.6 5.0
Hs20562732 29.6 5.1
Hs4759030 28.9 8.6
> YLR106c
Length=4910
Score = 96.3 bits (238), Expect = 4e-20, Method: Composition-based stats.
Identities = 50/93 (53%), Positives = 62/93 (66%), Gaps = 5/93 (5%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLT----EG 172
++E +FEW D LV AV KG WL+L +A+LCS SVLDRLN+LLE GSLL+ E
Sbjct 2185 TKEASVKFEWFDGMLVKAVEKGHWLILDNANLCSPSVLDRLNSLLEIDGSLLINECSQED 2244
Query 173 GAPREVKRHPNFRILLTAEAANTHLLSAALRNR 205
G PR +K HPNFR+ LT + LS A+RNR
Sbjct 2245 GQPRVLKPHPNFRLFLTMDPKYGE-LSRAMRNR 2276
Score = 58.5 bits (140), Expect = 9e-09, Method: Composition-based stats.
Identities = 30/77 (38%), Positives = 46/77 (59%), Gaps = 2/77 (2%)
Query 113 AAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE--DGGSLLLT 170
A + S E F +V+ +LV +R G+WLLL + +L +A L+ ++ LL D S+LL+
Sbjct 798 AQSSSIENSFVFNFVEGSLVKTIRAGEWLLLDEVNLATADTLESISDLLTEPDSRSILLS 857
Query 171 EGGAPREVKRHPNFRIL 187
E G +K HP+FRI
Sbjct 858 EKGDAEPIKAHPDFRIF 874
Score = 58.2 bits (139), Expect = 1e-08, Method: Composition-based stats.
Identities = 36/86 (41%), Positives = 46/86 (53%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRHP 182
FEW D L+ A+R G + LL + L SVL+RLN++LE SLLL E G+ V
Sbjct 1472 FEWSDGPLIQAMRTGNFFLLDEISLADDSVLERLNSVLEPERSLLLAEQGSSDSLVTASE 1531
Query 183 NFRILLTAEAANTH---LLSAALRNR 205
NF+ T + LS ALRNR
Sbjct 1532 NFQFFATMNPGGDYGKKELSPALRNR 1557
Score = 49.7 bits (117), Expect = 4e-06, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 46/91 (50%), Gaps = 6/91 (6%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
++ + + + LV A+RKG W++L + +L VL+ LN LL+D L + E V
Sbjct 1125 DDTGKLSFKEGVLVEALRKGYWIVLDELNLAPTDVLEALNRLLDDNRELFIPE--TQEVV 1182
Query 179 KRHPNFRILLTAEAANTH----LLSAALRNR 205
HP+F + T + +LS A RNR
Sbjct 1183 HPHPDFLLFATQNPPGIYGGRKILSRAFRNR 1213
Score = 47.8 bits (112), Expect = 1e-05, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 2/69 (2%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
E +F W D+ + A++KG+W+LL + +L S SVL+ LNA L+ G + E
Sbjct 1790 ERSGEFLWHDAPFLRAMKKGEWVLLDEMNLASQSVLEGLNACLDHRGEAYIPE--LDISF 1847
Query 179 KRHPNFRIL 187
HPNF +
Sbjct 1848 SCHPNFLVF 1856
Score = 35.8 bits (81), Expect = 0.071, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 36/80 (45%), Gaps = 3/80 (3%)
Query 117 SQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPR 176
S ++ FEW L AV++G+W+L+ D VL L +LLE + + G
Sbjct 358 SGDKPGTFEWRAGVLATAVKEGRWVLIEDIDKAPTDVLSILLSLLEKRELTIPSRG---E 414
Query 177 EVKRHPNFRILLTAEAANTH 196
VK F+++ T H
Sbjct 415 TVKAANGFQLISTVRINEDH 434
> SPCC737.08
Length=4717
Score = 87.0 bits (214), Expect = 3e-17, Method: Composition-based stats.
Identities = 45/96 (46%), Positives = 59/96 (61%), Gaps = 5/96 (5%)
Query 114 AACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLL---- 169
A S +FEW D L+ AV +G W +L +A+LCS +VLDRLN+LLE G L++
Sbjct 2004 AEFSASPAGRFEWFDGYLLKAVEEGHWFVLDNANLCSPAVLDRLNSLLEHKGVLIVNEKT 2063
Query 170 TEGGAPREVKRHPNFRILLTAEAANTHLLSAALRNR 205
TE G P+ +K HPNFR+ LT LS A+RNR
Sbjct 2064 TEDGHPKTIKPHPNFRLFLTVNPVYGE-LSRAMRNR 2098
Score = 55.1 bits (131), Expect = 1e-07, Method: Composition-based stats.
Identities = 28/68 (41%), Positives = 39/68 (57%), Gaps = 2/68 (2%)
Query 120 EMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVK 179
E QF W D+ +AA+R G W+LL + +L S SVL+ LNA L+ + E + K
Sbjct 1612 EGGQFAWRDAPFLAAMRNGHWVLLDELNLASQSVLEGLNACLDHRNEAYIPE--LDKVFK 1669
Query 180 RHPNFRIL 187
HPNFR+
Sbjct 1670 AHPNFRVF 1677
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 36/97 (37%), Positives = 47/97 (48%), Gaps = 22/97 (22%)
Query 122 SQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRH 181
S FEW D ALV A+++G + LL + L SVL+RLN++LE +L L E H
Sbjct 1294 SLFEWHDGALVTAMKQGDFFLLDEISLADDSVLERLNSVLELSRTLTLVE---------H 1344
Query 182 PNFRILLTAEAANTHL-------------LSAALRNR 205
N + LTA+ LS ALRNR
Sbjct 1345 SNAAVSLTAKDGFAFFATMNPGGDYGKKELSPALRNR 1381
Score = 48.9 bits (115), Expect = 6e-06, Method: Composition-based stats.
Identities = 40/149 (26%), Positives = 63/149 (42%), Gaps = 20/149 (13%)
Query 74 VQRRRQQRAWRLLFQGCTLAGLAHVLESVSSSSAAAAAAAAACSQEEMSQF--------- 124
+ R R + +L QG T +G ++E V+ + ++ ++
Sbjct 886 IARACSTRMFPILIQGPTSSGKTSMIEYVAKKTGHKFVRINNHEHTDLQEYIGTYVTDDN 945
Query 125 ---EWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRH 181
+ + LV A+R G W++L + +L VL+ LN LL+D L + E VK H
Sbjct 946 GSLSFREGVLVEALRNGYWIVLDELNLAPTDVLEALNRLLDDNRELFIPETQV--LVKPH 1003
Query 182 PNFRILLTAE-----AANTHLLSAALRNR 205
P F + T A H LS A RNR
Sbjct 1004 PEFMLFATQNPPGVYAGRKH-LSRAFRNR 1031
Score = 47.0 bits (110), Expect = 3e-05, Method: Composition-based stats.
Identities = 26/65 (40%), Positives = 37/65 (56%), Gaps = 1/65 (1%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGS-LLLTEGGAPREVKRHP 182
F +V+ ALV AVR G W+LL + +L S L+ + LL S +LL+E G + H
Sbjct 625 FSFVEGALVKAVRSGHWVLLDEINLASLETLEPIGQLLSSYESGILLSERGDITPITPHK 684
Query 183 NFRIL 187
NFR+
Sbjct 685 NFRLF 689
Score = 30.4 bits (67), Expect = 2.5, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLE 162
+FEW L AV G+W+L + + VL L LLE
Sbjct 205 EFEWQPGVLTQAVITGKWILFTNIEHAPSEVLSVLLPLLE 244
> 7303557
Length=4865
Score = 82.0 bits (201), Expect = 9e-16, Method: Compositional matrix adjust.
Identities = 48/125 (38%), Positives = 65/125 (52%), Gaps = 6/125 (4%)
Query 88 QGCTLAGLAHVLESVSSSSAAAAAAAAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDAH 147
Q C + L + E S A S FEWVDS +V +++ GQ++LL +
Sbjct 2217 QNCQVRALQQLAEIKSQLLLLQTYIKTADSLNTGGSFEWVDSQIVTSLKCGQYILLEHVN 2276
Query 148 LCSASVLDRLNALLEDGGSLLLTEGGAPRE-----VKRHPNFRILLTAEAANTHLLSAAL 202
LCS++VLDRLN + E GSLL+ E G + V + NFR LT + N LS A+
Sbjct 2277 LCSSAVLDRLNPVFEPNGSLLIAEKGISAQDSAEVVYKSTNFRAFLTVDPKNGE-LSRAM 2335
Query 203 RNRCC 207
RNRC
Sbjct 2336 RNRCV 2340
Score = 58.5 bits (140), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 48/86 (55%), Gaps = 5/86 (5%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
F ++ +LV + G W+LL + +L SA L+ L+ +LE GS++L E G VKRHP+
Sbjct 851 FAFIPGSLVNCISNGDWVLLDEINLASAETLECLSTILEPEGSVVLLERGDFTPVKRHPD 910
Query 184 FRILLTAEAANTHL----LSAALRNR 205
FRI NT + L +RNR
Sbjct 911 FRI-FACMNPNTDIGKKDLPVGIRNR 935
Score = 52.0 bits (123), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 36/108 (33%), Positives = 55/108 (50%), Gaps = 16/108 (14%)
Query 116 CSQEEMSQ-FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGA 174
C +E +Q FEW D L+ A+++G + + + L SVL+RLN +LE ++LL E G
Sbjct 1465 CRGKESTQLFEWADGPLIYAMQEGSYFMADEISLAEDSVLERLNCILEPERTVLLAEKGG 1524
Query 175 PREVKRHPNFRILLTAEAANTHL-------------LSAALRNRCCLV 209
E+ P+F ++ A++ L LS ALRNR V
Sbjct 1525 ISELDASPDF--VVQAKSGFQFLATMNPGGDFGKKELSPALRNRFTEV 1570
Score = 50.1 bits (118), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 36/137 (26%), Positives = 60/137 (43%), Gaps = 18/137 (13%)
Query 85 LLFQGCTLAGLAHVLESVSSSSAAAAAAAAACSQEEMSQF------------EWVDSALV 132
+L QG T AG +++ V+ S ++ ++ + + LV
Sbjct 1112 ILLQGPTSAGKTSLIDYVARRSGNRCLRINNHEHTDLQEYIGTYAADLDGKLTFREGVLV 1171
Query 133 AAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTAEA 192
A+R G W++L + +L S +L+ LN +L+D L + E VK HPNF + T
Sbjct 1172 QAMRHGFWIILDELNLASTDILEALNRVLDDNRELYIAETQTL--VKAHPNFMLFATQNP 1229
Query 193 ANTH----LLSAALRNR 205
+ LS A +NR
Sbjct 1230 PGLYGGRKTLSRAFKNR 1246
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 37/61 (60%), Gaps = 1/61 (1%)
Query 112 AAAACSQEEMSQFEWVDSALVAAVR-KGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLT 170
+ A ++ ++ F W D L+AA++ K W+LL + +L SVL+ LNA+L+ G + +
Sbjct 1813 SNTAATERQIRSFVWRDGPLLAALKAKNTWILLDELNLAPQSVLEGLNAVLDHRGEVYIP 1872
Query 171 E 171
E
Sbjct 1873 E 1873
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+F W+ L AV G WLLL D + L++LLE L+ G VK P
Sbjct 425 EFVWLPGVLTQAVMHGYWLLLEDLDAATQDTYTILSSLLE---RQCLSVPGFRDSVKIEP 481
Query 183 NFRILLTA 190
F++ +T
Sbjct 482 GFQLFVTV 489
> CE17878
Length=2030
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 55/84 (65%), Gaps = 2/84 (2%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRH 181
+FEW DS V A G WLL+ D +LCSA+VLDRLN+ LE GG L++ E E ++ H
Sbjct 1776 RFEWTDSVFVDAYLHGDWLLIEDVNLCSAAVLDRLNSCLESGGKLVVAERQNSYEPLEPH 1835
Query 182 PNFRILLTAEAANTHLLSAALRNR 205
P+FR+ L+ + A +S A+RNR
Sbjct 1836 PDFRVFLSMD-ARVGEISRAMRNR 1858
Score = 58.5 bits (140), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 34/86 (39%), Positives = 50/86 (58%), Gaps = 4/86 (4%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE-VKRHP 182
FEW D +++A+R G LL+ + L SVL+RLN L E+ +LLL++ G E V+ P
Sbjct 1064 FEWSDGVVISAMRDGVPLLVDEISLAEDSVLERLNPLFEEDRALLLSDAGTDTEVVESKP 1123
Query 183 NFRILLTAEAANTH---LLSAALRNR 205
F+++ T + LS ALRNR
Sbjct 1124 GFQMIATMNPGGDYGKKELSKALRNR 1149
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 34/64 (53%), Gaps = 0/64 (0%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
F W D ++ A++KG+W+LL + +L S SVL+ LNA + L + E E+ N
Sbjct 1407 FRWEDGPVLKAIKKGEWILLDEMNLASQSVLEGLNACFDHRRVLYIAELNREFEIPATSN 1466
Query 184 FRIL 187
R
Sbjct 1467 CRFF 1470
> At1g67120
Length=5138
Score = 78.6 bits (192), Expect = 9e-15, Method: Composition-based stats.
Identities = 37/92 (40%), Positives = 61/92 (66%), Gaps = 5/92 (5%)
Query 118 QEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTE----GG 173
+++ ++FEWV L+ A+ KG+W++L++A+LC+ +VLDR+N+L+E GS+ + E G
Sbjct 2036 KKQSTKFEWVTGMLIKAIEKGEWVVLKNANLCNPTVLDRINSLVEPCGSITINECGIVNG 2095
Query 174 APREVKRHPNFRILLTAEAANTHLLSAALRNR 205
P V HPNFR+ L+ +S A+RNR
Sbjct 2096 EPVTVVPHPNFRLFLSVNPKFGE-VSRAMRNR 2126
Score = 54.7 bits (130), Expect = 1e-07, Method: Composition-based stats.
Identities = 36/93 (38%), Positives = 49/93 (52%), Gaps = 5/93 (5%)
Query 118 QEEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAP-- 175
Q+ + F W D LV A+R G +L+ + L SVL+R+N++LE L L E G P
Sbjct 1304 QKWRAIFVWQDGPLVEAMRAGNIVLVDEISLADDSVLERMNSVLETDRKLSLAEKGGPVL 1363
Query 176 REVKRHPNFRILLTAEAANTH---LLSAALRNR 205
EV H +F +L T + LS ALRNR
Sbjct 1364 EEVVAHEDFFVLATMNPGGDYGKKELSPALRNR 1396
Score = 51.6 bits (122), Expect = 1e-06, Method: Composition-based stats.
Identities = 30/82 (36%), Positives = 43/82 (52%), Gaps = 6/82 (7%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
+ ALV AVR G W++L + +L + VL+ LN LL+D L + E + HPNF +
Sbjct 1017 EGALVKAVRGGHWIVLDELNLAPSDVLEALNRLLDDNRELFVPE--LSETISAHPNFMLF 1074
Query 188 LTAEAANTH----LLSAALRNR 205
T + +LS A RNR
Sbjct 1075 ATQNPPTLYGGRKILSRAFRNR 1096
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 27/65 (41%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLED-GGSLLLTEGGAPREVKRHP 182
F +V+ A V A+R+G W+LL + +L +L RL +LE GSL L E G + RH
Sbjct 712 FTFVEGAFVTALREGHWVLLDEVNLAPPEILGRLIGVLEGVRGSLCLAERGDVMGIPRHL 771
Query 183 NFRIL 187
NFR+
Sbjct 772 NFRLF 776
Score = 34.7 bits (78), Expect = 0.15, Method: Composition-based stats.
Identities = 21/83 (25%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
++ +F W +L A+ G W++L D + V L++LL S L ++G R
Sbjct 309 DQPGEFRWQPGSLTQAIMNGFWVVLEDIDKAPSDVPLVLSSLLGGSCSFLTSQGEEIRIA 368
Query 179 KRHPNFRILLTAEAANTHLLSAA 201
+ F + T E + +H+ A
Sbjct 369 ETFQLFSTISTPECSVSHIRDAG 391
> ECU08g1900
Length=2832
Score = 57.8 bits (138), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 48/83 (57%), Gaps = 2/83 (2%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPN 183
FEW DSA+V++V++G+ ++ SV DRLN+L E +L + E G V +P
Sbjct 1517 FEWCDSAVVSSVKEGKMVVFAGVEFVEKSVFDRLNSLFESERTLNVYERGVDTNVAVNPL 1576
Query 184 FRILLTAEAANTHLLSAALRNRC 206
R++L A+ +LS AL +RC
Sbjct 1577 TRLVLVAKEPG--MLSPALLDRC 1597
Score = 57.0 bits (136), Expect = 2e-08, Method: Composition-based stats.
Identities = 37/89 (41%), Positives = 48/89 (53%), Gaps = 5/89 (5%)
Query 120 EMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVK 179
E S+ W D LV A++ G L+ + +L SVL+RLN++LE +L +TE G EV
Sbjct 1002 ENSKIVWKDGPLVKAMKCGDAFLIDEINLAEDSVLERLNSVLESQRTLYITETG--EEVV 1059
Query 180 RHPNFRILLTAEAA---NTHLLSAALRNR 205
H FRIL T LS ALRNR
Sbjct 1060 AHDGFRILATMNPGGDFGKRELSPALRNR 1088
Score = 50.1 bits (118), Expect = 3e-06, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 4/83 (4%)
Query 125 EWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNF 184
E+ + ALV A+R+G W++L + +L + VL+ LN LL+D + + E V H NF
Sbjct 709 EFKEGALVGAMRRGDWVILDELNLACSDVLEVLNRLLDDNRQIYIPE--TDEVVVPHENF 766
Query 185 RILLTAEA--ANTHLLSAALRNR 205
RI T LS A RNR
Sbjct 767 RIFATQNIGYGGRKGLSKAFRNR 789
Score = 38.1 bits (87), Expect = 0.013, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 39/67 (58%), Gaps = 4/67 (5%)
Query 123 QFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHP 182
+ +++S L+ ++R G W++L + +LC+ SV++ LN++L+ L + E V H
Sbjct 1286 EIRFMESELLHSLRDGGWIILDEINLCTQSVIEGLNSVLDYRRKLFIPE----VTVDVHE 1341
Query 183 NFRILLT 189
+ RI T
Sbjct 1342 DTRIFAT 1348
Score = 34.3 bits (77), Expect = 0.18, Method: Composition-based stats.
Identities = 19/66 (28%), Positives = 33/66 (50%), Gaps = 1/66 (1%)
Query 119 EEMSQFEWVDSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREV 178
E+ + F + L + G+WLLL + +L S L+ ++ +L + L E G R +
Sbjct 422 EKRAHFVYKRGILTECMVNGEWLLLDEINLSSEETLNLIDGVL-GKKEIFLYESGDLRPL 480
Query 179 KRHPNF 184
+ HP F
Sbjct 481 RIHPGF 486
> CE20667
Length=1302
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 39/77 (50%), Gaps = 15/77 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE---------- 177
DSALV A R GQ L++ +A V+ L +LL D G+L+L +G R
Sbjct 723 DSALVKAARSGQILVIDEADKAPLHVIAILKSLL-DTGTLVLGDGRTLRPASSFTEADKT 781
Query 178 ----VKRHPNFRILLTA 190
V HPNFRI++ A
Sbjct 782 NDRIVPIHPNFRIIMLA 798
> CE20666
Length=1767
Score = 40.8 bits (94), Expect = 0.002, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 39/77 (50%), Gaps = 15/77 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPRE---------- 177
DSALV A R GQ L++ +A V+ L +LL D G+L+L +G R
Sbjct 723 DSALVKAARSGQILVIDEADKAPLHVIAILKSLL-DTGTLVLGDGRTLRPASSFTEADKT 781
Query 178 ----VKRHPNFRILLTA 190
V HPNFRI++ A
Sbjct 782 NDRIVPIHPNFRIIMLA 798
> CE23997
Length=4568
Score = 34.7 bits (78), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 27/83 (32%), Positives = 39/83 (46%), Gaps = 22/83 (26%)
Query 124 FEWVDSALVAAVRKGQWLLLRDAHLCS---ASVLDRLNALLEDGGSLLLTEGGAPREVKR 180
F DSAL A + G+W+LL++ HL A + RL+++ K
Sbjct 4025 FNQADSALGTATKSGRWVLLKNVHLAPSWLAQLEKRLHSM------------------KP 4066
Query 181 HPNFRILLTAEAANTHLLSAALR 203
H FR+ LTAE + L S+ LR
Sbjct 4067 HAQFRLFLTAE-IHPKLPSSILR 4088
> Hs4758176
Length=798
Score = 34.7 bits (78), Expect = 0.16, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 32/66 (48%), Gaps = 13/66 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
++AL A +KG W++L++ HL A L L LE+ HP FR+
Sbjct 255 EAALDLAAKKGHWVILQNIHLV-AKWLSTLEKKLEEHSE------------NSHPEFRVF 301
Query 188 LTAEAA 193
++AE A
Sbjct 302 MSAEPA 307
> Hs13876382
Length=4486
Score = 34.3 bits (77), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 32/66 (48%), Gaps = 13/66 (19%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRIL 187
++AL A +KG W++L++ HL A L L LE+ HP FR+
Sbjct 3943 EAALDLAAKKGHWVILQNIHLV-AKWLSTLEKKLEEHSE------------NSHPEFRVF 3989
Query 188 LTAEAA 193
++AE A
Sbjct 3990 MSAEPA 3995
> 7300687
Length=4472
Score = 31.2 bits (69), Expect = 1.5, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 35/71 (49%), Gaps = 19/71 (26%)
Query 128 DSALVAAVRKGQWLLLRDAHLCSA--SVLD-RLNALLEDGGSLLLTEGGAPREVKRHPNF 184
++A+ A + G W++L++ HL VL+ +L ED HP++
Sbjct 3928 EAAMDTAAKHGHWVVLQNIHLVRKWLPVLEKKLEYYAEDS----------------HPDY 3971
Query 185 RILLTAEAANT 195
R+ L+AE A+T
Sbjct 3972 RMFLSAEPAST 3982
> 7298401
Length=4010
Score = 31.2 bits (69), Expect = 1.7, Method: Composition-based stats.
Identities = 22/71 (30%), Positives = 37/71 (52%), Gaps = 13/71 (18%)
Query 134 AVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLTAEAA 193
V+ G W++L++ HL +AS + L + E+ LL + HP+FR+ LT+ A
Sbjct 3451 GVKMGNWVVLQNCHL-AASFMPLLEKICEN----LLPDA-------THPDFRLWLTSYPA 3498
Query 194 NTHLLSAALRN 204
+ H L+N
Sbjct 3499 D-HFPVVVLQN 3508
> Hs16507235
Length=4523
Score = 30.4 bits (67), Expect = 2.5, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 13/66 (19%)
Query 130 ALVAAVRKGQWLLLRDAHLCSASVLDRLNALLEDGGSLLLTEGGAPREVKRHPNFRILLT 189
AL A + G W++L++ HL A L L LLE ++G H ++R+ ++
Sbjct 3982 ALEKASKGGHWVILQNVHLV-AKWLGTLEKLLER-----FSQGS-------HRDYRVFMS 4028
Query 190 AEAANT 195
AE+A T
Sbjct 4029 AESAPT 4034
> Hs22049728
Length=2274
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 32/62 (51%), Gaps = 7/62 (11%)
Query 90 CTLAGL---AHVLESVSSSSAAAAAAAAACSQEEMSQFEWVDSALVAAVRKGQWLLLRDA 146
C++ G HV E +++ + A S E +Q D A+ AV+ G+W++L++
Sbjct 1672 CSVPGYDASGHV-EDLAAEQNTQITSIAIGSAEGFNQ---ADKAINTAVKSGRWVMLKNV 1727
Query 147 HL 148
HL
Sbjct 1728 HL 1729
> 7296871
Length=1260
Score = 29.6 bits (65), Expect = 4.5, Method: Composition-based stats.
Identities = 22/76 (28%), Positives = 38/76 (50%), Gaps = 4/76 (5%)
Query 8 EAAACTTEDAPRAAPEAQQQQQQLQQRLAE--ETAALLKACEKAATTLCSGLSGDNSPAA 65
E A T +DA A EA++Q+ +L+++ A + +LLK K L+ ++P +
Sbjct 278 EGAGLTPQDA--VAQEAEKQKDELKKQRANYVKKVSLLKKEMKVLKDQREDLAAGDAPPS 335
Query 66 ATTADFAAVQRRRQQR 81
TT +F R Q +
Sbjct 336 PTTKNFIEENDRLQMQ 351
> Hs4504097
Length=453
Score = 29.6 bits (65), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 24/55 (43%), Gaps = 6/55 (10%)
Query 85 LLFQGCTLAGLAHVLESVSSSSAA------AAAAAAACSQEEMSQFEWVDSALVA 133
LF+ C+ A L HVL A A + C + + F WV SALVA
Sbjct 113 FLFEACSYATLLHVLTLSFERYIAICHPFRYKAVSGPCQVKLLIGFVWVTSALVA 167
> Hs20562732
Length=269
Score = 29.6 bits (65), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 33/65 (50%), Gaps = 6/65 (9%)
Query 14 TEDAPRAAP-EAQQQQQQLQQRLAEETAALLKACEKAATTLCSGLSGDNSPAAATTADFA 72
TED P A P E ++ + L+QRLA E ++A E++ L S S +P A
Sbjct 146 TEDPPDALPREGKRGSEMLEQRLARE----IRALERSKEQLLSE-SEPPAPGRLVRAKLR 200
Query 73 AVQRR 77
V+RR
Sbjct 201 EVERR 205
> Hs4759030
Length=1208
Score = 28.9 bits (63), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 37 EETAALLKACEKAATTLCSGLSGDNSPAAATTADFAAVQRRRQQRAW 83
E AALL+ C AA SG + A A A + +RRR QRA+
Sbjct 714 ERIAALLRTCLHAAWVPGSGGRAPKTTAEAYHAGMCSRERRRVQRAF 760
Lambda K H
0.317 0.124 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3754464630
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40