bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3849_orf1
Length=107
Score E
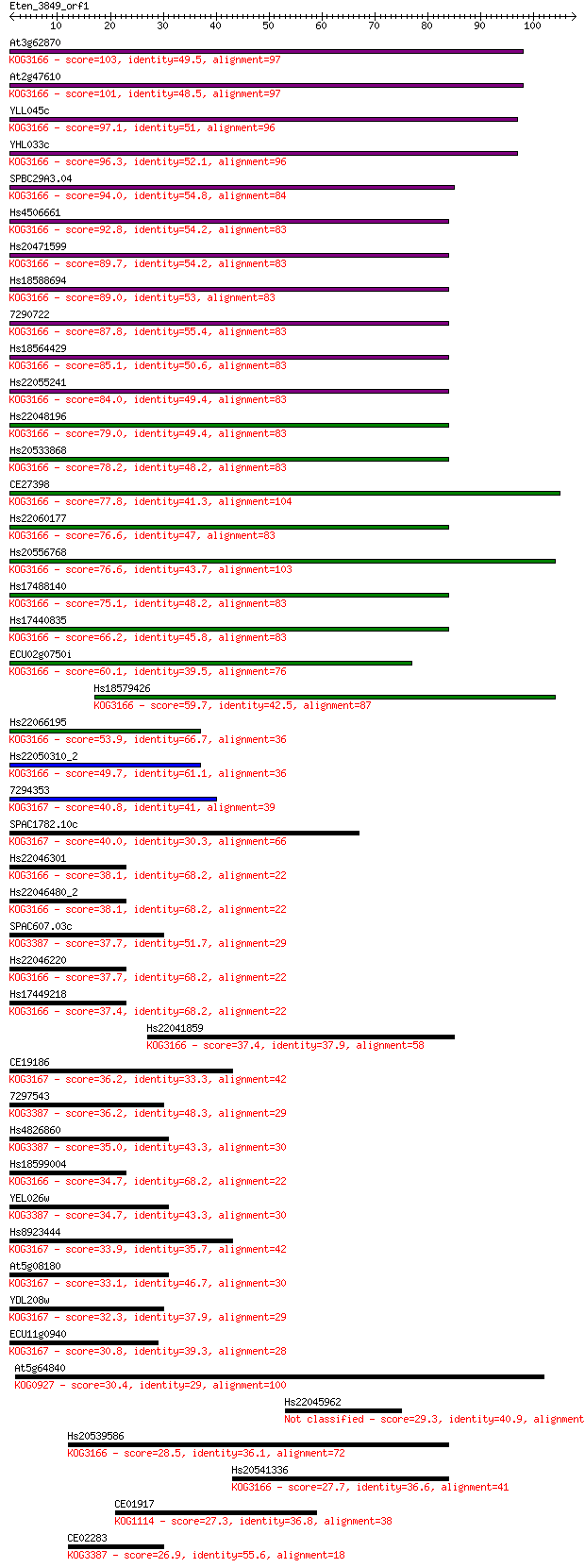
Sequences producing significant alignments: (Bits) Value
At3g62870 103 9e-23
At2g47610 101 3e-22
YLL045c 97.1 7e-21
YHL033c 96.3 1e-20
SPBC29A3.04 94.0 6e-20
Hs4506661 92.8 1e-19
Hs20471599 89.7 1e-18
Hs18588694 89.0 2e-18
7290722 87.8 4e-18
Hs18564429 85.1 3e-17
Hs22055241 84.0 6e-17
Hs22048196 79.0 2e-15
Hs20533868 78.2 4e-15
CE27398 77.8 4e-15
Hs22060177 76.6 9e-15
Hs20556768 76.6 1e-14
Hs17488140 75.1 3e-14
Hs17440835 66.2 1e-11
ECU02g0750i 60.1 1e-09
Hs18579426 59.7 1e-09
Hs22066195 53.9 7e-08
Hs22050310_2 49.7 1e-06
7294353 40.8 6e-04
SPAC1782.10c 40.0 0.001
Hs22046301 38.1 0.004
Hs22046480_2 38.1 0.004
SPAC607.03c 37.7 0.005
Hs22046220 37.7 0.005
Hs17449218 37.4 0.007
Hs22041859 37.4 0.007
CE19186 36.2 0.014
7297543 36.2 0.017
Hs4826860 35.0 0.030
Hs18599004 34.7 0.044
YEL026w 34.7 0.045
Hs8923444 33.9 0.072
At5g08180 33.1 0.11
YDL208w 32.3 0.21
ECU11g0940 30.8 0.66
At5g64840 30.4 0.82
Hs22045962 29.3 1.7
Hs20539586 28.5 2.8
Hs20541336 27.7 5.6
CE01917 27.3 7.2
CE02283 26.9 8.8
> At3g62870
Length=256
Score = 103 bits (256), Expect = 9e-23, Method: Compositional matrix adjust.
Identities = 48/98 (48%), Positives = 67/98 (68%), Gaps = 1/98 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV WLPALCRK +VPYCIVKGK+RLG +VH+KT A + + V+ ED+ E +
Sbjct 153 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTAAALCLTTVKNEDKLEFSKILEAI 212
Query 61 RARFKDN-SDLRRRWGGGHLGIKSQHAQEKKQKLIDAE 97
+A F D + R++WGGG +G KSQ + K+++I E
Sbjct 213 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 250
> At2g47610
Length=257
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 47/98 (47%), Positives = 67/98 (68%), Gaps = 1/98 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV WLPALCRK +VPYCIVKGK+RLG +VH+KT + + + V+ ED+ E +
Sbjct 154 PIELVVWLPALCRKMEVPYCIVKGKSRLGAVVHQKTASCLCLTTVKNEDKLEFSKILEAI 213
Query 61 RARFKDN-SDLRRRWGGGHLGIKSQHAQEKKQKLIDAE 97
+A F D + R++WGGG +G KSQ + K+++I E
Sbjct 214 KANFNDKYEEYRKKWGGGIMGSKSQAKTKAKERVIAKE 251
> YLL045c
Length=256
Score = 97.1 bits (240), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 49/97 (50%), Positives = 64/97 (65%), Gaps = 1/97 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPY I+KGKARLG LV++KT AV A+ VR ED+A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIIKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDA 96
A F D D +++ WGGG LG K+Q +K+ K D+
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAKTSDS 255
> YHL033c
Length=256
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 50/97 (51%), Positives = 64/97 (65%), Gaps = 1/97 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPY IVKGKARLG LV++KT AV A+ VR ED+A L
Sbjct 159 PIELVVFLPALCKKMGVPYAIVKGKARLGTLVNQKTSAVAALTEVRAEDEAALAKLVSTI 218
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDA 96
A F D D +++ WGGG LG K+Q +K+ K D+
Sbjct 219 DANFADKYDEVKKHWGGGILGNKAQAKMDKRAKNSDS 255
> SPBC29A3.04
Length=259
Score = 94.0 bits (232), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 46/85 (54%), Positives = 59/85 (69%), Gaps = 1/85 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC+K VPY IVK KARLG ++H+KT AV+AV VR+ED+ EL +
Sbjct 159 PIELVVFLPALCKKMGVPYAIVKNKARLGTVIHQKTAAVLAVTEVREEDKNELASIVSAV 218
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQ 84
A F D RR+WGGG +G K+Q
Sbjct 219 DANFSAKYDESRRKWGGGIMGGKTQ 243
> Hs4506661
Length=266
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALCRK VPYCI+KGKARLG+LVH+KT +A V ED+ L +
Sbjct 163 PIELVVFLPALCRKMGVPYCIIKGKARLGRLVHRKTCTTVAFTQVNSEDKGALAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> Hs20471599
Length=259
Score = 89.7 bits (221), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 45/84 (53%), Positives = 55/84 (65%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI LV +LPALCRK VPYCI+KGKARLG+LVH+KT +A+ V ED+ L +
Sbjct 163 PIGLVIFLPALCRKMGVPYCIIKGKARLGRLVHRKTCTTVALTQVHLEDKGGLAKLVEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> Hs18588694
Length=266
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 44/84 (52%), Positives = 54/84 (64%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +L ALCRK VPYCI+KGKARLG+LVH+KT +A V ED+ L +
Sbjct 163 PIELVVFLAALCRKMGVPYCIIKGKARLGRLVHRKTCTTVAFTQVNSEDKGTLAKLMEAI 222
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR WGG LG KS
Sbjct 223 RTNYNDRYDEIRRHWGGNVLGPKS 246
> 7290722
Length=271
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 46/87 (52%), Positives = 58/87 (66%), Gaps = 7/87 (8%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAE----LEAQ 56
P++LV +LPALCRK VPYCIVKGKARLG+LV +KT +A+ V D+A LEA
Sbjct 168 PLELVLFLPALCRKMGVPYCIVKGKARLGRLVRRKTCTTLALTTVDNNDKANFGKVLEAV 227
Query 57 CKNYRARFKDNSDLRRRWGGGHLGIKS 83
N+ R + ++RR WGGG LG KS
Sbjct 228 KTNFNER---HEEIRRHWGGGILGSKS 251
> Hs18564429
Length=294
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 42/84 (50%), Positives = 54/84 (64%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LPALC K VPYCI+KG+ARLG+LVH+KT +A V ED+ L
Sbjct 191 PIELVVFLPALCHKMRVPYCIIKGEARLGRLVHRKTCTTVAFIQVNSEDKGALAKLVGAI 250
Query 61 RARFKDN-SDLRRRWGGGHLGIKS 83
R + D +++RR WGG LG KS
Sbjct 251 RTDYNDRYNEIRRHWGGNVLGPKS 274
> Hs22055241
Length=260
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI++V +LPALCRK VPY I+KGKARLG LVH+KT +A V E + L +
Sbjct 157 PIEVVVFLPALCRKMGVPYSIIKGKARLGHLVHRKTCTTVAFTQVNSEGKGALAKLVETI 216
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +R WGG LG KS
Sbjct 217 RTNYNDRYDEIRHHWGGNVLGPKS 240
> Hs22048196
Length=220
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 41/84 (48%), Positives = 51/84 (60%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI LV +LPALC K +VPYC++K KARLG LVH+KT A V E++ L +
Sbjct 117 PIKLVVFLPALCHKMEVPYCVIKRKARLGCLVHRKTCTNAAFTQVNSENKGALAKLVEAI 176
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D SD + R WGG LG KS
Sbjct 177 RTNYNDRSDEICRHWGGKALGPKS 200
> Hs20533868
Length=229
Score = 78.2 bits (191), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 40/84 (47%), Positives = 52/84 (61%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+L +LPALCRK V CI+K KARLG+LVH+KT + V ED+ L + +
Sbjct 126 PIELAVFLPALCRKMGVSCCIIKRKARLGRLVHRKTCTTVVFTQVNLEDKGALAKRVEAI 185
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R +K+ SD +R WGG LG KS
Sbjct 186 RTNYKNRSDEIRHHWGGNVLGPKS 209
> CE27398
Length=265
Score = 77.8 bits (190), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 62/105 (59%), Gaps = 4/105 (3%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P+++V LPALCRK +VPY I+KGKA LG +V +KT A +A+ V ED++ L +
Sbjct 164 PLEIVLHLPALCRKYNVPYAIIKGKASLGTVVRRKTTAAVALVDVNPEDKSALNKLVETV 223
Query 61 RARFKD-NSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGGL 104
F + + ++R+ WGGG + KS KK K+ A R G L
Sbjct 224 NNNFSERHEEIRKHWGGGVMSAKS---DAKKLKIERARARDLGKL 265
> Hs22060177
Length=223
Score = 76.6 bits (187), Expect = 9e-15, Method: Compositional matrix adjust.
Identities = 39/84 (46%), Positives = 48/84 (57%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+LV +LP LC K P+CI+KGKARLG LVH+KT +A V ED+ L +
Sbjct 120 PIELVVFLPVLCHKMGFPHCIIKGKARLGCLVHRKTSTTVAFTEVNSEDKGALAKLVEAI 179
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R D D + WGG LG KS
Sbjct 180 RTNSSDRYDEIHCHWGGNVLGPKS 203
> Hs20556768
Length=431
Score = 76.6 bits (187), Expect = 1e-14, Method: Composition-based stats.
Identities = 45/104 (43%), Positives = 56/104 (53%), Gaps = 1/104 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
P +LV +LPALC K VPYCI+KGKARLG LVH+KT + V ED+ L +
Sbjct 328 PTELVGFLPALCCKMRVPYCIIKGKARLGHLVHRKTCTTVTFTQVNSEDKGALAKLVEAI 387
Query 61 RARFKDNSD-LRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKKGG 103
R + D D + WGG LG KS K +K E+ K G
Sbjct 388 RTNYDDRYDEIHCHWGGNVLGHKSVAHVTKLEKAKARELATKLG 431
> Hs17488140
Length=384
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 40/84 (47%), Positives = 50/84 (59%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI+ V +LPALCRK VPYCI+KGKARLG+LVH++ +A V ED+ L
Sbjct 243 PIEPVVFLPALCRKMGVPYCIIKGKARLGRLVHREICTTVAFTQVNSEDKGALAKLVAAI 302
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R + D D +RR G LG KS
Sbjct 303 RTDYNDRYDEIRRHRGSNVLGPKS 326
> Hs17440835
Length=253
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/84 (45%), Positives = 47/84 (55%), Gaps = 1/84 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI LV +LPALC K VPYC++KGKARLG+LVH+KT +A ED+ L +
Sbjct 150 PIKLVVFLPALCCKIWVPYCLIKGKARLGRLVHRKTCTTVAFTQFNWEDRGVLMKLMEAI 209
Query 61 RARFKDNSD-LRRRWGGGHLGIKS 83
R D D + W LG KS
Sbjct 210 RTNDNDRYDEICHHWESNVLGPKS 233
> ECU02g0750i
Length=193
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/77 (38%), Positives = 43/77 (55%), Gaps = 1/77 (1%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNY 60
PI++V +LP LCRK V Y IV+ LG+LV+ KT + + + VR ED+ + +
Sbjct 107 PIEVVIFLPTLCRKMGVSYAIVENSTLLGKLVNLKTTSCVCLCDVRPEDEGSFKEMLRTA 166
Query 61 RARFKDNSDLR-RRWGG 76
A F DN + WGG
Sbjct 167 DAIFLDNYETHLSTWGG 183
> Hs18579426
Length=90
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 37/88 (42%), Positives = 46/88 (52%), Gaps = 1/88 (1%)
Query 17 VPYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNYRARFKDNSD-LRRRWG 75
VPYCI+KGKARLG+LVH+KT +A V ED+ L + R D D + WG
Sbjct 3 VPYCIIKGKARLGRLVHRKTCTTVAFTQVNLEDKGALAKLVEGIRTNDNDRYDEICCHWG 62
Query 76 GGHLGIKSQHAQEKKQKLIDAEMRKKGG 103
G LG KS K +K E+ K G
Sbjct 63 GNILGPKSVACIAKLEKAKAKELATKLG 90
> Hs22066195
Length=139
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 24/36 (66%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKT 36
P++LV +LP L K VPYCI+KGKARLG LVH+KT
Sbjct 104 PVELVVFLPVLHCKMGVPYCIIKGKARLGHLVHRKT 139
> Hs22050310_2
Length=132
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/36 (61%), Positives = 29/36 (80%), Gaps = 0/36 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKT 36
PI+L+ +LPAL K +P+CI+KGKARL LVH+KT
Sbjct 96 PIELIVFLPALRHKMRIPHCIIKGKARLECLVHRKT 131
> 7294353
Length=160
Score = 40.8 bits (94), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 24/39 (61%), Gaps = 0/39 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKKTGAV 39
P+D++C LPA+C +K +PY +A LG + K G V
Sbjct 94 PVDIMCHLPAVCEEKGIPYTYTPSRADLGAAMGVKRGTV 132
> SPAC1782.10c
Length=154
Score = 40.0 bits (92), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 2/68 (2%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLVHKK--TGAVIAVDAVRKEDQAELEAQCK 58
P+D++ +P LC +VPY K LG+ + K T V+ V +K+D +++E +
Sbjct 81 PMDVISHIPVLCEDNNVPYLYTVSKELLGEASNTKRPTSCVMIVPGGKKKDMSKVEEYKE 140
Query 59 NYRARFKD 66
+Y K+
Sbjct 141 SYEEIIKE 148
> Hs22046301
Length=202
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIV 22
PI+LV +LPALC K VPYCI+
Sbjct 181 PIELVVFLPALCCKLGVPYCII 202
> Hs22046480_2
Length=181
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIV 22
PI+LV +LPALC K VPYCI+
Sbjct 160 PIELVVFLPALCCKLGVPYCII 181
> SPAC607.03c
Length=125
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
PI+++ LP LC K+VPY V KA LG
Sbjct 59 PIEILLHLPLLCEDKNVPYVFVPSKAALG 87
> Hs22046220
Length=124
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIV 22
PI+LV +LPALC K VPYCI+
Sbjct 103 PIELVVFLPALCCKLGVPYCII 124
> Hs17449218
Length=139
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 18/22 (81%), Gaps = 0/22 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIV 22
PI+LV +LPALC K VPYCI+
Sbjct 118 PIELVVFLPALCCKLGVPYCII 139
> Hs22041859
Length=108
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 1/59 (1%)
Query 27 RLGQLVHKKTGAVIAVDAVRKEDQAELEAQCKNYRARFKDNSD-LRRRWGGGHLGIKSQ 84
RLG+LV++KT +A V ED+ L + R +KD D + WGG L KS
Sbjct 31 RLGRLVYRKTCTTVAFAWVNSEDKGALAKLVEAIRTNYKDRHDEICCHWGGNVLASKSM 89
> CE19186
Length=163
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 26/43 (60%), Gaps = 1/43 (2%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAV 42
PID+ +P +C +K++PY + + +LG V H++ +I V
Sbjct 96 PIDVYSHIPGICEEKEIPYVYIPSREQLGLAVGHRRPSILIFV 138
> 7297543
Length=160
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 19/29 (65%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
PI+++ LP LC K+VPY V+ K LG
Sbjct 94 PIEILLHLPLLCEDKNVPYVFVRSKQALG 122
> Hs4826860
Length=128
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
P++++ LP LC K+VPY V+ K LG+
Sbjct 62 PLEIILHLPLLCEDKNVPYVFVRSKQALGR 91
> Hs18599004
Length=160
Score = 34.7 bits (78), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 15/22 (68%), Positives = 17/22 (77%), Gaps = 0/22 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIV 22
PI+LV +LPAL K VPYCIV
Sbjct 139 PIELVVFLPALYCKLSVPYCIV 160
> YEL026w
Length=126
Score = 34.7 bits (78), Expect = 0.045, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
PI+++ LP LC K+VPY V + LG+
Sbjct 60 PIEILLHLPLLCEDKNVPYVFVPSRVALGR 89
> Hs8923444
Length=153
Score = 33.9 bits (76), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 23/43 (53%), Gaps = 1/43 (2%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQLV-HKKTGAVIAV 42
PI++ C LP +C +++PY + K LG K+ VI V
Sbjct 87 PIEVYCHLPVMCEDRNLPYVYIPSKTDLGAAAGSKRPTCVIMV 129
> At5g08180
Length=156
Score = 33.1 bits (74), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLGQ 30
PID++ LP LC + VPY V K L Q
Sbjct 78 PIDVITHLPILCEEAGVPYVYVPSKEDLAQ 107
> YDL208w
Length=173
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 11/29 (37%), Positives = 15/29 (51%), Gaps = 0/29 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARLG 29
P D++ +P LC VPY + K LG
Sbjct 100 PADVISHIPVLCEDHSVPYIFIPSKQDLG 128
> ECU11g0940
Length=109
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 1 PIDLVCWLPALCRKKDVPYCIVKGKARL 28
P+DL+ LPALC + Y V+ K+ +
Sbjct 57 PMDLITHLPALCEDRGTKYVFVRSKSSI 84
> At5g64840
Length=692
Score = 30.4 bits (67), Expect = 0.82, Method: Composition-based stats.
Identities = 29/112 (25%), Positives = 51/112 (45%), Gaps = 12/112 (10%)
Query 2 IDLVCWLPALCRKKDVPYCIVK-GKARLGQLVHKKTGAVIAVDAVRKEDQAE-------- 52
+D + WL +K+DVP I+ +A L QL K + V + + ++
Sbjct 292 LDTIEWLEGYLQKQDVPMVIISHDRAFLDQLCTKIVETEMGVSRTFEGNYSQYVISKAEW 351
Query 53 LEAQCKNYRARFKD---NSDLRRRWGGGHLGIKSQHAQEKKQKLIDAEMRKK 101
+E Q + + KD DL R G G ++ A++K +KL + E+ +K
Sbjct 352 IETQNAAWEKQQKDIDSTKDLIARLGAGANSGRASTAEKKLEKLQEQELIEK 403
> Hs22045962
Length=205
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 9/22 (40%), Positives = 14/22 (63%), Gaps = 0/22 (0%)
Query 53 LEAQCKNYRARFKDNSDLRRRW 74
++A C ++R F N DL+ RW
Sbjct 4 IKANCTDFREEFASNVDLKNRW 25
> Hs20539586
Length=199
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 26/77 (33%), Positives = 36/77 (46%), Gaps = 9/77 (11%)
Query 12 CRKKDV----PYCIVKGKARLGQLVHKKTGAVIAVDAVRKEDQAEL-EAQCKNYRARFKD 66
C K D+ P + G + LV K ++ V+ KE A+L EA NY R+
Sbjct 107 CSKGDISTRRPLVLQAGVNTVTTLVENKKAQLV-VNLEDKEALAKLVEAIRTNYNNRY-- 163
Query 67 NSDLRRRWGGGHLGIKS 83
++ R WGG LG KS
Sbjct 164 -DEICRHWGGNVLGPKS 179
> Hs20541336
Length=180
Score = 27.7 bits (60), Expect = 5.6, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 19/42 (45%), Gaps = 1/42 (2%)
Query 43 DAVRKEDQAELEAQCKNYRARFKDNSD-LRRRWGGGHLGIKS 83
V ED+ L + R + D D + R WGG LG KS
Sbjct 119 SGVNSEDKRALAKLVEAIRTNYNDKYDEIHRHWGGSVLGPKS 160
> CE01917
Length=1374
Score = 27.3 bits (59), Expect = 7.2, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 21 IVKGKARLGQLVHKKTGAVIAVDAVRKEDQAELEAQCK 58
+++G L LVH K G I+VD A L +CK
Sbjct 955 VLQGMKELPLLVHVKLGNKISVDLAASASDATLGKECK 992
> CE02283
Length=128
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 10/18 (55%), Positives = 12/18 (66%), Gaps = 0/18 (0%)
Query 12 CRKKDVPYCIVKGKARLG 29
C K+VPY V+ KA LG
Sbjct 73 CEDKNVPYVFVRSKAALG 90
Lambda K H
0.321 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164169380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40