bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3882_orf1
Length=238
Score E
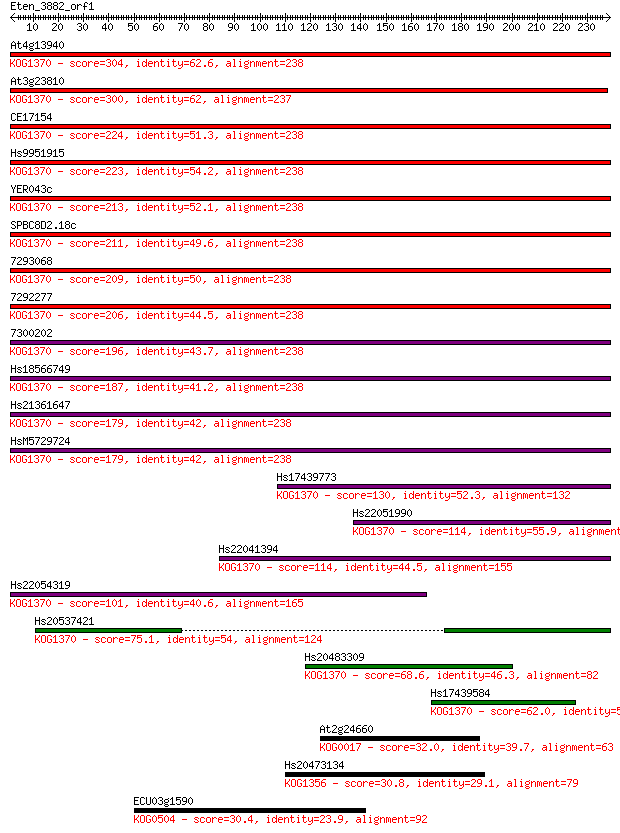
Sequences producing significant alignments: (Bits) Value
At4g13940 304 1e-82
At3g23810 300 2e-81
CE17154 224 2e-58
Hs9951915 223 4e-58
YER043c 213 2e-55
SPBC8D2.18c 211 7e-55
7293068 209 5e-54
7292277 206 4e-53
7300202 196 4e-50
Hs18566749 187 1e-47
Hs21361647 179 5e-45
HsM5729724 179 6e-45
Hs17439773 130 2e-30
Hs22051990 114 1e-25
Hs22041394 114 2e-25
Hs22054319 101 1e-21
Hs20537421 75.1 1e-13
Hs20483309 68.6 1e-11
Hs17439584 62.0 1e-09
At2g24660 32.0 1.1
Hs20473134 30.8 2.5
ECU03g1590 30.4 3.1
> At4g13940
Length=485
Score = 304 bits (778), Expect = 1e-82, Method: Compositional matrix adjust.
Identities = 149/238 (62%), Positives = 186/238 (78%), Gaps = 1/238 (0%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA + SA VFAWKGET++EYWWC+E+AL W GPDL+VDDGGD T
Sbjct 86 CNIFSTQDHAAAAIARD-SAAVFAWKGETLQEYWWCTERALDWGPGGGPDLIVDDGGDAT 144
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
+LIHEGV AEE++E +G +P+ ++ + E V +++ + P+K+ ++ + GVSE
Sbjct 145 LLIHEGVKAEEIFEKTGQVPDPTSTDNPEFQIVLSIIKEGLQVDPKKYHKMKERLVGVSE 204
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV RL + LLFPA+NVND VTKSKFDN+YGCRHSLPD +MRATDVM+ GK+
Sbjct 205 ETTTGVKRLYQMQQNGTLLFPAINVNDSVTKSKFDNLYGCRHSLPDGLMRATDVMIAGKV 264
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AV+CGYGDVGKGCAAA++ G RVI++EIDPICALQA MEG VL L+ VV AD+FV
Sbjct 265 AVICGYGDVGKGCAAAMKTAGARVIVTEIDPICALQALMEGLQVLTLEDVVSEADIFV 322
> At3g23810
Length=485
Score = 300 bits (767), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 147/237 (62%), Positives = 183/237 (77%), Gaps = 1/237 (0%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA + SA VFAWKGET++EYWWC+E+AL W GPDL+VDDGGD T
Sbjct 86 CNIFSTQDHAAAAIARD-SAAVFAWKGETLQEYWWCTERALDWGPGGGPDLIVDDGGDAT 144
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
+LIHEGV AEE++ +G P+ ++ + E V +++ + P+K+ ++ + GVSE
Sbjct 145 LLIHEGVKAEEIFAKNGTFPDPTSTDNPEFQIVLSIIKDGLQVDPKKYHKMKERLVGVSE 204
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV RL + LLFPA+NVND VTKSKFDN+YGCRHSLPD +MRATDVM+ GK+
Sbjct 205 ETTTGVKRLYQMQETGALLFPAINVNDSVTKSKFDNLYGCRHSLPDGLMRATDVMIAGKV 264
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
AV+CGYGDVGKGCAAA++ G RVI++EIDPICALQA MEG VL L+ VV AD+F
Sbjct 265 AVICGYGDVGKGCAAAMKTAGARVIVTEIDPICALQALMEGLQVLTLEDVVSEADIF 321
> CE17154
Length=437
Score = 224 bits (570), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 122/238 (51%), Positives = 146/238 (61%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ + + D ++++DDGGD+T
Sbjct 80 CNIFSTQDHAAAAIAQTG-VPVYAWKGETDEEYEWCIEQTIVFKDGQPLNMILDDGGDLT 138
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+H K+ Q AG+RG+SE
Sbjct 139 NLVHA-----------------------------------------KYPQYLAGIRGLSE 157
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + A L PA+NVND VTKSKFDN+YG R SLPD I RATDVML GK+
Sbjct 158 ETTTGVHNLAKMLAKGDLKVPAINVNDSVTKSKFDNLYGIRESLPDGIKRATDVMLAGKV 217
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
AVV GYGDVGKG AA+L+ G RVI++EIDPI ALQAAMEG+ V L+ A++ V
Sbjct 218 AVVAGYGDVGKGSAASLKAFGSRVIVTEIDPINALQAAMEGYEVTTLEEAAPKANIIV 275
> Hs9951915
Length=432
Score = 223 bits (567), Expect = 4e-58, Method: Compositional matrix adjust.
Identities = 129/239 (53%), Positives = 150/239 (62%), Gaps = 45/239 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGP-DLLVDDGGDI 59
CNIFSTQDHAAAAIA G V+AWKGET EEY WC EQ L + D GP ++++DDGGD+
Sbjct 79 CNIFSTQDHAAAAIAKAG-IPVYAWKGETDEEYLWCIEQTLYFKD--GPLNMILDDGGDL 135
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T LIH K+ QL G+RG+S
Sbjct 136 TNLIHT-----------------------------------------KYPQLLPGIRGIS 154
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L + A +L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK
Sbjct 155 EETTTGVHNLYKMMANGILKVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGK 214
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV GYGDVGKGCA ALRG G RVII+EIDPI ALQAAMEG+ V +D ++FV
Sbjct 215 VAVVAGYGDVGKGCAQALRGFGARVIITEIDPINALQAAMEGYEVTTMDEACQEGNIFV 273
> YER043c
Length=449
Score = 213 bits (543), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 124/239 (51%), Positives = 146/239 (61%), Gaps = 43/239 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQAL-TWPDCAGPDLLVDDGGDI 59
CNI+STQDHAAAAIAA G VFAWKGET EEY WC EQ L + D +L++DDGGD+
Sbjct 80 CNIYSTQDHAAAAIAASG-VPVFAWKGETEEEYLWCIEQQLFAFKDNKKLNLILDDGGDL 138
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T L+HE K ++ G+S
Sbjct 139 TTLVHE-----------------------------------------KHPEMLEDCFGLS 157
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L R L PA+NVND VTKSKFDN+YGCR SL D I RATDVML GK
Sbjct 158 EETTTGVHHLYRMVKEGKLKVPAINVNDSVTKSKFDNLYGCRESLVDGIKRATDVMLAGK 217
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV GYGDVGKGCAAALRG G RV+++EIDPI ALQAAMEG+ V+ ++ + +FV
Sbjct 218 VAVVAGYGDVGKGCAAALRGMGARVLVTEIDPINALQAAMEGYQVVTMEDASHIGQVFV 276
> SPBC8D2.18c
Length=433
Score = 211 bits (538), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 118/239 (49%), Positives = 148/239 (61%), Gaps = 43/239 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQAL-TWPDCAGPDLLVDDGGDI 59
CNI+STQDHAAAAIAA G VFAWKGET EEY WC EQ L ++P ++++DDGGD+
Sbjct 78 CNIYSTQDHAAAAIAATG-VPVFAWKGETEEEYLWCIEQQLKSFPSGKPLNMILDDGGDL 136
Query 60 TMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVS 119
T L+HE + +L +RG+S
Sbjct 137 TALVHE-----------------------------------------RHPELLVDIRGIS 155
Query 120 EETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGK 179
EETTTGV L + N L PA+NVND VTKSKFDN++GC+ SL D I RATDVM+ GK
Sbjct 156 EETTTGVHNLYKMFKENKLKVPAINVNDSVTKSKFDNLFGCKESLVDGIKRATDVMIAGK 215
Query 180 LAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
+AVV G+GDVGKGC+ +LR QG RVI++E+DPI ALQAAM+GF V ++ V +FV
Sbjct 216 VAVVAGFGDVGKGCSTSLRSQGARVIVTEVDPINALQAAMDGFEVTTMEEAVKEGQIFV 274
> 7293068
Length=432
Score = 209 bits (531), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 119/238 (50%), Positives = 145/238 (60%), Gaps = 42/238 (17%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQD+AAAAIAA G V+AWKGET EEY WC EQ L +PD ++++DDGGD+T
Sbjct 78 CNIFSTQDNAAAAIAATG-VPVYAWKGETDEEYMWCIEQTLVFPDGQPLNMILDDGGDLT 136
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+HE K+ Q ++G+SE
Sbjct 137 NLVHE-----------------------------------------KFPQYLKNIKGLSE 155
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
ETTTGV L + L PA+NVND VTKSKFDN+YGCR SL D I RATDVM+ GK+
Sbjct 156 ETTTGVHNLYKMFKEGRLGVPAINVNDSVTKSKFDNLYGCRESLIDGIKRATDVMIAGKV 215
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
V GYGDVGKGCA AL+G G RVI++E+DPI ALQAAMEG+ V ++ A +FV
Sbjct 216 CCVAGYGDVGKGCAQALKGFGGRVIVTEVDPINALQAAMEGYEVTTMEEASKEASIFV 273
> 7292277
Length=521
Score = 206 bits (523), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 106/238 (44%), Positives = 145/238 (60%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A G +FAW+GET E++WWC ++ + + P++++DDGGD T
Sbjct 168 CNIYSTQNEVAAALAESG-IPIFAWRGETEEDFWWCIDRCVNAENWQ-PNMILDDGGDAT 225
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+ +K+ + V+G+ E
Sbjct 226 HLML-----------------------------------------KKYPTMFKLVKGIVE 244
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK+KFDN+Y C+ S+ D++ R+TDVM GGK
Sbjct 245 ESVTGVHRLYQLSKAGKLTVPAMNVNDSVTKTKFDNLYSCKESILDSLKRSTDVMFGGKQ 304
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYGDVGKGCA AL+GQGC V I+EIDPICALQA+M+GF V+ L+ V+ D+ V
Sbjct 305 VVVCGYGDVGKGCAQALKGQGCIVYITEIDPICALQASMDGFRVVKLNEVIRNVDIVV 362
> 7300202
Length=504
Score = 196 bits (497), Expect = 4e-50, Method: Compositional matrix adjust.
Identities = 104/238 (43%), Positives = 141/238 (59%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A G +FAW+GET EE+WWC ++A+ + D P+L++DDGGD T
Sbjct 140 CNIYSTQNAVAAALAEAG-IPIFAWRGETEEEFWWCLDRAI-YSDGWQPNLILDDGGDAT 197
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L+ +K+ +RG+ E
Sbjct 198 HLM-----------------------------------------LKKYPDYFKAIRGIVE 216
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + L PA+NVND VTK+KFD Y CR S+ D++ R TD+M GGK
Sbjct 217 ESVTGVHRLYMLSKGGKLTVPAINVNDSVTKNKFDTFYTCRDSILDSLKRTTDIMFGGKQ 276
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
V+CGYGDVGKGCA +L+GQGC V ++E+DPICALQAAM+GF V+ L+ V+ D+ V
Sbjct 277 VVICGYGDVGKGCAQSLKGQGCIVYVTEVDPICALQAAMDGFRVVRLNEVIRTVDVVV 334
> Hs18566749
Length=611
Score = 187 bits (475), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 98/241 (40%), Positives = 138/241 (57%), Gaps = 49/241 (20%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALT---WPDCAGPDLLVDDGG 57
CNI+ST + AAA+A G VFAWKGE+ +++WWC ++ + W P++++DDGG
Sbjct 258 CNIYSTLNEVAAALAESGFP-VFAWKGESEDDFWWCIDRCVNVEGW----QPNMILDDGG 312
Query 58 DITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRG 117
D+T I+ +K+ + ++G
Sbjct 313 DLTHWIY-----------------------------------------KKYPNMFKKIKG 331
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
+ EE+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TD+M G
Sbjct 332 IVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDMMFG 391
Query 178 GKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLF 237
GK VVCGYG+VGKGC AAL+ G V ++EIDPICALQA M+GF ++ L+ V+ D+
Sbjct 392 GKQVVVCGYGEVGKGCCAALKAMGSIVYVTEIDPICALQACMDGFRLVKLNEVIRQVDIV 451
Query 238 V 238
+
Sbjct 452 I 452
> Hs21361647
Length=530
Score = 179 bits (453), Expect = 5e-45, Method: Compositional matrix adjust.
Identities = 100/238 (42%), Positives = 137/238 (57%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AAA+A VFAWKGE+ +++WWC ++ + D ++++DDGGD+T
Sbjct 177 CNIYSTQNEVAAALAEA-GVAVFAWKGESEDDFWWCIDRCVNM-DGWQANMILDDGGDLT 234
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + +RG+ E
Sbjct 235 HWVY-----------------------------------------KKYPNVFKKIRGIVE 253
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM GGK
Sbjct 254 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQ 313
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC AAL+ G V I+EIDPICALQA M+GF V+ L+ V+ D+ +
Sbjct 314 VVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGFRVVKLNEVIRQVDVVI 371
> HsM5729724
Length=500
Score = 179 bits (453), Expect = 6e-45, Method: Compositional matrix adjust.
Identities = 100/238 (42%), Positives = 136/238 (57%), Gaps = 43/238 (18%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNI+STQ+ AA AE VFAWKGE+ +++WWC ++ + D ++++DDGGD+T
Sbjct 147 CNIYSTQN-EVAAALAEAGVAVFAWKGESEDDFWWCIDRCVNM-DGWQANMILDDGGDLT 204
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
++ +K+ + +RG+ E
Sbjct 205 HWVY-----------------------------------------KKYPNVFKKIRGIVE 223
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKL 180
E+ TGV RL + + A L PAMNVND VTK KFDN+Y CR S+ D + R TDVM GGK
Sbjct 224 ESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQ 283
Query 181 AVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VVCGYG+VGKGC AAL+ G V I+EIDPICALQA M+GF V+ L+ V+ D+ +
Sbjct 284 VVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGFRVVKLNEVIRQVDVVI 341
> Hs17439773
Length=413
Score = 130 bits (328), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 69/132 (52%), Positives = 86/132 (65%), Gaps = 6/132 (4%)
Query 107 KWRQLAAGVRGVSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPD 166
++ QL RG+S+ETT GV L + +L PA++VND TKSKFDN+YGC
Sbjct 129 EYSQLLLSFRGISKETTIGVHNLYKMMVNGILKVPAIDVNDSATKSKFDNLYGC------ 182
Query 167 AIMRATDVMLGGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLP 226
I +ATDVM+ GK+AVV GYGD GK CA ALRG VII++ DPI ALQAAMEG+ V
Sbjct 183 GIKQATDVMIAGKVAVVAGYGDKGKSCAQALRGFRASVIITKTDPIKALQAAMEGYEVTT 242
Query 227 LDAVVGVADLFV 238
+D D+FV
Sbjct 243 MDEACQEGDVFV 254
> Hs22051990
Length=648
Score = 114 bits (286), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 57/102 (55%), Positives = 74/102 (72%), Gaps = 0/102 (0%)
Query 137 LLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKLAVVCGYGDVGKGCAAA 196
+L PA+NVND +TKS+F+ +YGC SL D I AT VM+ GK+A+V GYG+VGKGCA A
Sbjct 483 ILKVPAINVNDSLTKSEFNKLYGCWESLIDGIKWATVVMIAGKVAMVAGYGNVGKGCAQA 542
Query 197 LRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
L G G VII++IDPI ALQAAMEG+ V +D ++F+
Sbjct 543 LWGFGAHVIITKIDPINALQAAMEGYEVTTMDEACQEGNIFI 584
> Hs22041394
Length=283
Score = 114 bits (285), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 69/157 (43%), Positives = 94/157 (59%), Gaps = 10/157 (6%)
Query 84 AAADEEAAAVQQLLRRSVAAAPRKWRQLAA--GVRGVSEETTTGVLRLLRRAAANLLLFP 141
+ D A + +L + + ++ R +A G RG+S+ET TGV L + A +L P
Sbjct 45 STQDHAALPLPRLAFQCMPGREKQTRSTSAPVGHRGISQETLTGVHNLHKMMANGILKVP 104
Query 142 AMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLGGKLAVVCGYGDVGKGCAAALRGQG 201
A+NVND VTKSK + SL D IM+AT+V+ K+ V+ GYG+VGK CA ALRG
Sbjct 105 AINVNDSVTKSK-------QASLTDGIMQATEVVTASKVVVIVGYGNVGKDCAQALRGFV 157
Query 202 CRVIISEIDPICALQAAMEGFGVLPLDAVVGVADLFV 238
VI++EIDPI ALQAAME + V +D D+FV
Sbjct 158 DHVIVTEIDPINALQAAMESYEVTTMDKACQ-EDIFV 193
> Hs22054319
Length=287
Score = 101 bits (251), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 67/165 (40%), Positives = 84/165 (50%), Gaps = 43/165 (26%)
Query 1 CNIFSTQDHAAAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGPDLLVDDGGDIT 60
CNIFSTQDHAAAAIA G V AWKGET E+Y WC++Q L + D ++++D G +T
Sbjct 24 CNIFSTQDHAAAAIAKAG-IPVNAWKGETEEKYLWCTKQTLYFKD-KLLNMILDITGGLT 81
Query 61 MLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWRQLAAGVRGVSE 120
L H K+ QL +G+R +SE
Sbjct 82 NLTH-----------------------------------------TKYPQLLSGIRDISE 100
Query 121 ETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLP 165
ET T V L + A +L +NVND VTKSKFDN+YGC SLP
Sbjct 101 ETMTEVHNLYKMMANGILKVSTINVNDSVTKSKFDNLYGCHQSLP 145
> Hs20537421
Length=215
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 38/66 (57%), Positives = 47/66 (71%), Gaps = 0/66 (0%)
Query 173 DVMLGGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGVLPLDAVVG 232
DVM+ K+AVV GYG VGKGCA AL+G G +II+E DPI ALQAAMEG+ V +D
Sbjct 94 DVMIASKVAVVAGYGGVGKGCAQALQGFGACIIITETDPISALQAAMEGYEVTTMDEACQ 153
Query 233 VADLFV 238
++FV
Sbjct 154 EGNIFV 159
Score = 56.6 bits (135), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 29/59 (49%), Positives = 37/59 (62%), Gaps = 3/59 (5%)
Query 11 AAAIAAEGSATVFAWKGETVEEYWWCSEQALTWPDCAGP-DLLVDDGGDITMLIHEGVH 68
A A+ AE VF WKG+ E Y WC EQ L + D GP ++++DDGGD+T LIH H
Sbjct 33 AVAVFAEAGMPVFTWKGKMKEGYPWCIEQTLYFKD--GPLNMILDDGGDLTNLIHTKWH 89
> Hs20483309
Length=395
Score = 68.6 bits (166), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 51/82 (62%), Gaps = 0/82 (0%)
Query 118 VSEETTTGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSLPDAIMRATDVMLG 177
V +ET GV +L + A +L PA+NVN+ VTKSKFDN+ GC+ SL D+I ATDVM+
Sbjct 138 VGDETMIGVHKLHKMMANGILKVPAINVNNSVTKSKFDNLSGCQESLIDSIKWATDVMIA 197
Query 178 GKLAVVCGYGDVGKGCAAALRG 199
K V+ D AA++G
Sbjct 198 SKAHVIIIKIDPINILQAAMKG 219
> Hs17439584
Length=215
Score = 62.0 bits (149), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/57 (54%), Positives = 40/57 (70%), Gaps = 0/57 (0%)
Query 168 IMRATDVMLGGKLAVVCGYGDVGKGCAAALRGQGCRVIISEIDPICALQAAMEGFGV 224
I R TDVM+ GK+A+V GYGD+GKGCA AL G +I++I PI LQ AM G+ +
Sbjct 104 IKRDTDVMIVGKVAMVAGYGDMGKGCAQALWDFGDPFVITKIGPINLLQTAMGGYEI 160
> At2g24660
Length=1156
Score = 32.0 bits (71), Expect = 1.1, Method: Composition-based stats.
Identities = 25/69 (36%), Positives = 30/69 (43%), Gaps = 6/69 (8%)
Query 124 TGVLRLLRRAAANLLLFPAMNVNDCVTKSKFDN-VY-----GCRHSLPDAIMRATDVMLG 177
T V LLR AAN M+VN+ D VY G RHS PD + R + G
Sbjct 727 TTVRTLLRLVAANQWEVYQMDVNNAFLHGDLDEEVYMKLPPGFRHSHPDKVCRLRKSLYG 786
Query 178 GKLAVVCGY 186
K A C +
Sbjct 787 LKQAPRCWF 795
> Hs20473134
Length=1341
Score = 30.8 bits (68), Expect = 2.5, Method: Composition-based stats.
Identities = 23/84 (27%), Positives = 39/84 (46%), Gaps = 6/84 (7%)
Query 110 QLAAGVRGVSEETTTGV-----LRLLRRAAANLLLFPAMNVNDCVTKSKFDNVYGCRHSL 164
QLA+ R VSE ++ G L+L ++ P +N + TKS+ + + S+
Sbjct 123 QLASKDR-VSERSSAGAHKTDCLKLAEAGETGRIILPNVNSDSVHTKSEKNFQAVSQGSV 181
Query 165 PDAIMRATDVMLGGKLAVVCGYGD 188
P ++M A + M K V+ D
Sbjct 182 PSSVMSAVNTMCNTKTDVITSAAD 205
> ECU03g1590
Length=450
Score = 30.4 bits (67), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 22/92 (23%), Positives = 44/92 (47%), Gaps = 5/92 (5%)
Query 50 DLLVDDGGDITMLIHEGVHAEEVYESSGVLPEVSAAADEEAAAVQQLLRRSVAAAPRKWR 109
+LL+ GG++TM+ EGV E++ S G ++ E A+ Q+L SV +++
Sbjct 326 ELLIRLGGNMTMVNREGVSTEDILISQGKTFDIKEVDVELQEALFQILGDSV-----EFK 380
Query 110 QLAAGVRGVSEETTTGVLRLLRRAAANLLLFP 141
+ + + + +++ R +LL P
Sbjct 381 HYLSIIDRIKYNCSHTIVKKNRFTITSLLNLP 412
Lambda K H
0.320 0.134 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4740636838
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40