bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3903_orf3
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
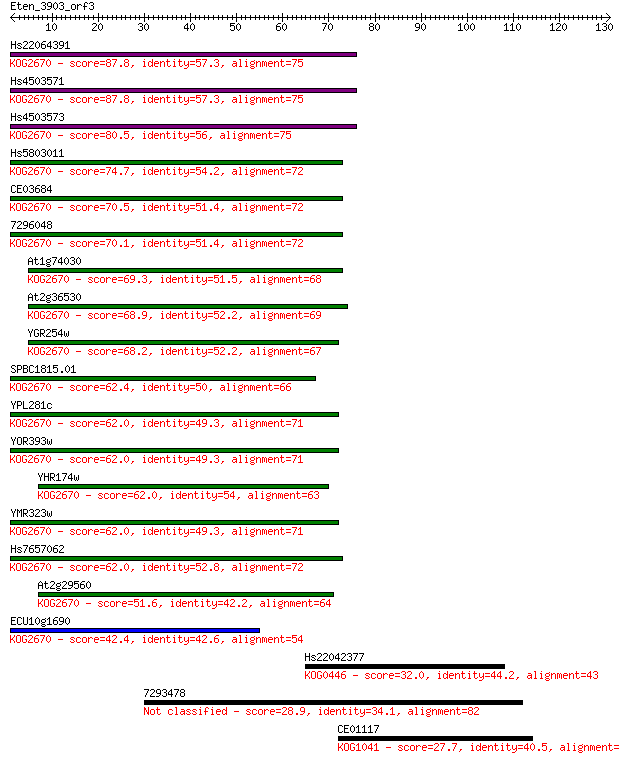
Hs22064391 87.8 4e-18
Hs4503571 87.8 5e-18
Hs4503573 80.5 7e-16
Hs5803011 74.7 4e-14
CE03684 70.5 7e-13
7296048 70.1 9e-13
At1g74030 69.3 2e-12
At2g36530 68.9 2e-12
YGR254w 68.2 3e-12
SPBC1815.01 62.4 2e-10
YPL281c 62.0 3e-10
YOR393w 62.0 3e-10
YHR174w 62.0 3e-10
YMR323w 62.0 3e-10
Hs7657062 62.0 3e-10
At2g29560 51.6 4e-07
ECU10g1690 42.4 2e-04
Hs22042377 32.0 0.34
7293478 28.9 2.3
CE01117 27.7 5.4
> Hs22064391
Length=377
Score = 87.8 bits (216), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 43/75 (57%), Positives = 49/75 (65%), Gaps = 0/75 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A + G GVMV H SGET D FI L G GQ K GAP RSE LAKY Q +I+ ELG+
Sbjct 303 AQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGS 362
Query 61 KPNFSGRGFKNPLPK 75
K F+GR F+NPL K
Sbjct 363 KAKFAGRNFRNPLAK 377
> Hs4503571
Length=434
Score = 87.8 bits (216), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 43/75 (57%), Positives = 49/75 (65%), Gaps = 0/75 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A + G GVMV H SGET D FI L G GQ K GAP RSE LAKY Q +I+ ELG+
Sbjct 360 AQANGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGS 419
Query 61 KPNFSGRGFKNPLPK 75
K F+GR F+NPL K
Sbjct 420 KAKFAGRNFRNPLAK 434
> Hs4503573
Length=434
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 42/75 (56%), Positives = 46/75 (61%), Gaps = 0/75 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A S G GVMV H SGET D FI L G GQ K GAP RSE LAKY Q +I+ LG
Sbjct 360 AQSNGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLMRIEEALGD 419
Query 61 KPNFSGRGFKNPLPK 75
K F+GR F+NP K
Sbjct 420 KAIFAGRKFRNPKAK 434
> Hs5803011
Length=434
Score = 74.7 bits (182), Expect = 4e-14, Method: Composition-based stats.
Identities = 39/72 (54%), Positives = 44/72 (61%), Gaps = 0/72 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A G GVMV H SGET D FI L G GQ K GAP RSE LAKY Q +I+ ELG
Sbjct 360 AQENGWGVMVSHRSGETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLMRIEEELGD 419
Query 61 KPNFSGRGFKNP 72
+ F+G F+NP
Sbjct 420 EARFAGHNFRNP 431
> CE03684
Length=434
Score = 70.5 bits (171), Expect = 7e-13, Method: Composition-based stats.
Identities = 37/72 (51%), Positives = 45/72 (62%), Gaps = 0/72 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
+ + G GVMV H SGET D FI L G + GQ K GAP RSE LAKY Q +I+ ELG
Sbjct 361 SRANGWGVMVSHRSGETEDTFIADLVVGLATGQIKTGAPCRSERLAKYNQLLRIEEELGA 420
Query 61 KPNFSGRGFKNP 72
++G F+NP
Sbjct 421 DAVYAGHNFRNP 432
> 7296048
Length=433
Score = 70.1 bits (170), Expect = 9e-13, Method: Composition-based stats.
Identities = 37/72 (51%), Positives = 43/72 (59%), Gaps = 0/72 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A G G MV H SGET D FI L G S GQ K GAP RSE LAKY Q +I+ E+G
Sbjct 361 AKKNGWGTMVSHRSGETEDSFIGDLVVGLSTGQIKTGAPCRSERLAKYNQILRIEEEIGA 420
Query 61 KPNFSGRGFKNP 72
F+G+ F+ P
Sbjct 421 GVKFAGKSFRKP 432
> At1g74030
Length=477
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/68 (51%), Positives = 43/68 (63%), Gaps = 1/68 (1%)
Query 5 GGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGTKPNF 64
G GVMV H SGET D FI L G + GQ K GAP RSE L+KY Q +I+ ELG +
Sbjct 411 GWGVMVSHRSGETEDNFIADLSVGLASGQIKTGAPCRSERLSKYNQLLRIEEELGN-VRY 469
Query 65 SGRGFKNP 72
+G F++P
Sbjct 470 AGEAFRSP 477
> At2g36530
Length=444
Score = 68.9 bits (167), Expect = 2e-12, Method: Composition-based stats.
Identities = 36/69 (52%), Positives = 44/69 (63%), Gaps = 0/69 (0%)
Query 5 GGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGTKPNF 64
G GVM H SGET D FI L G S GQ K GAP RSE LAKY Q +I+ ELG++ +
Sbjct 373 GWGVMTSHRSGETEDTFIADLAVGLSTGQIKTGAPCRSERLAKYNQLLRIEEELGSEAIY 432
Query 65 SGRGFKNPL 73
+G F+ P+
Sbjct 433 AGVNFRKPV 441
> YGR254w
Length=437
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/67 (52%), Positives = 40/67 (59%), Gaps = 0/67 (0%)
Query 5 GGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGTKPNF 64
G GVMV H SGET D FI L G GQ K GAP+RSE LAK Q +I+ ELG F
Sbjct 367 GWGVMVSHRSGETEDTFIADLVVGLRTGQIKTGAPARSERLAKLNQLLRIEEELGDNAVF 426
Query 65 SGRGFKN 71
+G F +
Sbjct 427 AGENFHH 433
> SPBC1815.01
Length=439
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 33/66 (50%), Positives = 43/66 (65%), Gaps = 0/66 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
++ G GVMV H SGET D FI L G GQ K+GAP RSE LAKY + +I+ ELG+
Sbjct 362 SYEAGWGVMVSHRSGETADTFISHLTVGIGAGQLKSGAPCRSERLAKYNELLRIEEELGS 421
Query 61 KPNFSG 66
+ ++G
Sbjct 422 EGVYAG 427
> YPL281c
Length=437
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A G GVM+ H SGET DPFI L G GQ K+GA SRSE LAKY + +I+ ELG
Sbjct 363 AFDAGWGVMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGD 422
Query 61 KPNFSGRGFKN 71
++G F +
Sbjct 423 DCIYAGHRFHD 433
> YOR393w
Length=437
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A G GVM+ H SGET DPFI L G GQ K+GA SRSE LAKY + +I+ ELG
Sbjct 363 AFDAGWGVMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGD 422
Query 61 KPNFSGRGFKN 71
++G F +
Sbjct 423 DCIYAGHRFHD 433
> YHR174w
Length=437
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 34/63 (53%), Positives = 39/63 (61%), Gaps = 0/63 (0%)
Query 7 GVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGTKPNFSG 66
GVMV H SGET D FI L G GQ K GAP+RSE LAK Q +I+ ELG K ++G
Sbjct 369 GVMVSHRSGETEDTFIADLVVGLRTGQIKTGAPARSERLAKLNQLLRIEEELGDKAVYAG 428
Query 67 RGF 69
F
Sbjct 429 ENF 431
> YMR323w
Length=437
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 35/71 (49%), Positives = 43/71 (60%), Gaps = 0/71 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGT 60
A G GVM+ H SGET DPFI L G GQ K+GA SRSE LAKY + +I+ ELG
Sbjct 363 AFDAGWGVMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGD 422
Query 61 KPNFSGRGFKN 71
++G F +
Sbjct 423 DCIYAGHRFHD 433
> Hs7657062
Length=458
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 38/75 (50%), Positives = 44/75 (58%), Gaps = 3/75 (4%)
Query 1 AHSKGGGVM-VFHP-SGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKR-E 57
A S G GVM V H SGET D F+ L G GQ K G RSE LAKY Q +I+ E
Sbjct 381 AQSNGWGVMPVSHRLSGETEDTFMADLVVGLCTGQIKTGPTCRSERLAKYNQLLRIEEAE 440
Query 58 LGTKPNFSGRGFKNP 72
G+K F+GR F+NP
Sbjct 441 AGSKARFAGRNFRNP 455
> At2g29560
Length=475
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 35/64 (54%), Gaps = 0/64 (0%)
Query 7 GVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKIKRELGTKPNFSG 66
GV+ H GET D FI L G + G K GAP R E KY Q +I+ ELG + ++G
Sbjct 409 GVVTSHRCGETEDSFISDLSVGLATGVIKAGAPCRGERTMKYNQLLRIEEELGDQAVYAG 468
Query 67 RGFK 70
+K
Sbjct 469 EDWK 472
> ECU10g1690
Length=412
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 23/54 (42%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 1 AHSKGGGVMVFHPSGETGDPFIFALGGGGSPGQKKNGAPSRSEGLAKYTQFFKI 54
A G +MV H SGET D FI L G K+GAP R E ++KY Q ++
Sbjct 356 ARKCGMKIMVSHRSGETDDHFISDLSVGVGAEYIKSGAPCRGERVSKYNQLLRL 409
> Hs22042377
Length=863
Score = 32.0 bits (71), Expect = 0.34, Method: Composition-based stats.
Identities = 19/45 (42%), Positives = 20/45 (44%), Gaps = 2/45 (4%)
Query 65 SGRGFKNPLPKKTGGGGAPPTPC--GPLKPPFDVPDQGGAPVDAP 107
SGRG +P GAPP P GPL P D GAP P
Sbjct 785 SGRGPAPAIPSPGPHSGAPPVPFRPGPLPPFPSSSDSFGAPPQVP 829
> 7293478
Length=1004
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 39/91 (42%), Gaps = 11/91 (12%)
Query 30 SPGQKKNGAPSRS-EGLAKYTQFFKIKRELGTKPNFSGRGFKNPL-----PKKTGGGGAP 83
SP + G P+ + + L K+ F + T NF + K P K +G GG+P
Sbjct 194 SPLAQVEGDPTSNIDDLKKHILF--LHNMTKTNSNFESKFVKFPSLQKDKAKTSGAGGSP 251
Query 84 PTPCGPLKP--PFDVPDQGGAP-VDAPRGLG 111
P P P +P + P P V AP G G
Sbjct 252 PNPKRPQRPIHQYSAPIAPPTPKVPAPDGGG 282
> CE01117
Length=1040
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 21/45 (46%), Gaps = 3/45 (6%)
Query 72 PLPKKTGGGGAPP-TPCGPLKPPFDVPDQGGAP--VDAPRGLGKL 113
P+ + G A P T G L PP DQG +D+PR L L
Sbjct 29 PITSRPASGQASPLTSNGSLSPPQYADDQGSVSYNLDSPRDLSPL 73
Lambda K H
0.313 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40