bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
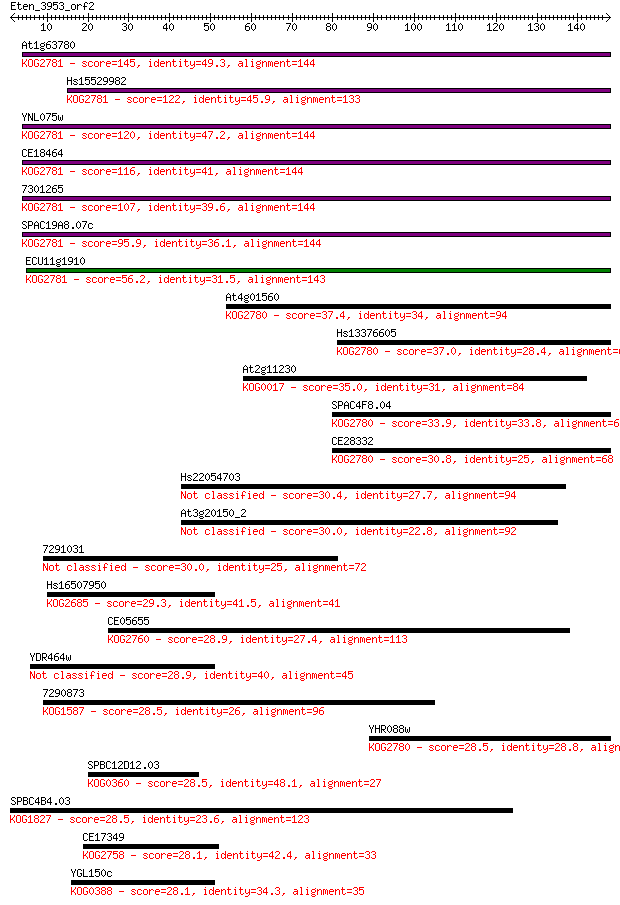
Query= Eten_3953_orf2
Length=147
Score E
Sequences producing significant alignments: (Bits) Value
At1g63780 145 3e-35
Hs15529982 122 2e-28
YNL075w 120 8e-28
CE18464 116 2e-26
7301265 107 7e-24
SPAC19A8.07c 95.9 2e-20
ECU11g1910 56.2 2e-08
At4g01560 37.4 0.009
Hs13376605 37.0 0.013
At2g11230 35.0 0.048
SPAC4F8.04 33.9 0.12
CE28332 30.8 0.91
Hs22054703 30.4 1.1
At3g20150_2 30.0 1.6
7291031 30.0 1.6
Hs16507950 29.3 3.1
CE05655 28.9 3.9
YDR464w 28.9 3.9
7290873 28.5 4.2
YHR088w 28.5 5.1
SPBC12D12.03 28.5 5.1
SPBC4B4.03 28.5 5.1
CE17349 28.1 6.6
YGL150c 28.1 6.9
> At1g63780
Length=294
Score = 145 bits (365), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 71/144 (49%), Positives = 100/144 (69%), Gaps = 5/144 (3%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
M RR RL+KEY++RKSLE ER++ E+K+ ++E L+ GKP+PT LR A +L Q
Sbjct 1 MQRRLVRLKKEYIYRKSLEGDERKVYEQKRLIREALQEGKPIPTELRNVEA-----KLRQ 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
+DL+D T P++H+DDEYA A DP I LTTSR+PS+ L +F KEL + PN +RIN
Sbjct 56 EIDLEDQNTAVPRSHIDDEYANATEADPKILLTTSRNPSAPLIRFTKELKFVFPNSQRIN 115
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG+ V+ E+++ RS TD++LV
Sbjct 116 RGSQVISEIIETARSHDFTDVILV 139
> Hs15529982
Length=291
Score = 122 bits (306), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 61/133 (45%), Positives = 87/133 (65%), Gaps = 5/133 (3%)
Query 15 YLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLDDPQTLA 74
YL+RK+ EE +R ERK++++ LE + +PT LR A A L L+ DD
Sbjct 12 YLYRKAREEAQRSAQERKERLRRALEENRLIPTELRREALA-----LQGSLEFDDAGGEG 66
Query 75 PKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQ 134
+HVDDEY +AGVEDP + +TTSR PSSRL+ F KEL L+ P +R+NRG + L++
Sbjct 67 VTSHVDDEYRWAGVEDPKVMITTSRDPSSRLKMFAKELKLVFPGAQRMNRGRHEVGALVR 126
Query 135 LCRSSGITDLLLV 147
C+++G+TDLL+V
Sbjct 127 ACKANGVTDLLVV 139
> YNL075w
Length=290
Score = 120 bits (302), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 68/149 (45%), Positives = 92/149 (61%), Gaps = 12/149 (8%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR AR R+EYL+RK+ E ++ QL +++Q +K+ L GKPLP L + R Q
Sbjct 1 MLRRQARERREYLYRKAQELQDSQLQQKRQIIKQALAQGKPLPKELAEDESLQKDFRYDQ 60
Query 64 VL----DLDDPQTLAPKTHVDDEYA-YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPN 118
L + DD Q VDDEYA +G+ DP I +TTSR PS+RL QF KE+ LL PN
Sbjct 61 SLKESEEADDLQ-------VDDEYAATSGIMDPRIIVTTSRDPSTRLSQFAKEIKLLFPN 113
Query 119 CRRINRGTIVLDELLQLCRSSGITDLLLV 147
R+NRG V+ L+ C+ SG TDL+++
Sbjct 114 AVRLNRGNYVMPNLVDACKKSGTTDLVVL 142
> CE18464
Length=292
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 59/144 (40%), Positives = 88/144 (61%), Gaps = 10/144 (6%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
M+RR RLR+E++FRKSLEEK++ L E++++++ LE + NLR A LA+
Sbjct 1 MIRRENRLRREFIFRKSLEEKQKSLEEKREKIRNALENNTKIDYNLRKDAI-----ELAK 55
Query 64 VLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRIN 123
D Q D EY +AG +DP I +TTSR PSSRL+ F KE+ L+ PN +RIN
Sbjct 56 GSDWGGQQY-----ETDSEYRWAGAQDPKIVITTSRDPSSRLKMFAKEMKLIFPNAQRIN 110
Query 124 RGTIVLDELLQLCRSSGITDLLLV 147
RG + +++Q ++ TDL++
Sbjct 111 RGHYDVKQVVQASKAQDSTDLIIF 134
> 7301265
Length=298
Score = 107 bits (268), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 57/145 (39%), Positives = 91/145 (62%), Gaps = 9/145 (6%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPT-NLRAAAAAAAAARLA 62
MLR+ AR R+EYL+ K+L E+ + + ++ V +++ K + + N++ + A + + A
Sbjct 1 MLRKQARQRREYLYNKALTERLKSKQKIQETVVKSINENKAIGSKNVKKSLTAYKSLKYA 60
Query 63 QVLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRI 122
+DD V+DEY YAG EDP I LTTS +PSSRL+ F+KEL L++PN +++
Sbjct 61 DE-GVDDRT-------VNDEYHYAGCEDPKIMLTTSHNPSSRLKMFMKELRLIIPNAQQM 112
Query 123 NRGTIVLDELLQLCRSSGITDLLLV 147
NRG L L+ CR++ +TD L+V
Sbjct 113 NRGNYQLTTLMHACRANNVTDFLIV 137
> SPAC19A8.07c
Length=289
Score = 95.9 bits (237), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 52/146 (35%), Positives = 93/146 (63%), Gaps = 8/146 (5%)
Query 4 MLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQ 63
MLRR R R++++++++ E +E +L E+++ +++ LE K L +L+ + +L +
Sbjct 1 MLRRAVRERRQFIYKRNQELQEAKLNEKRRALRKALEGNKELNKDLQEDS------QLQK 54
Query 64 VLDLDDPQTL--APKTHVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRR 121
D+ + +T++DDEY G +P + +TTSR PSSRL QF KE+ LL+PN R
Sbjct 55 DYKYDESRATQEETETNLDDEYHRLGEREPKVLVTTSREPSSRLAQFAKEVRLLIPNSYR 114
Query 122 INRGTIVLDELLQLCRSSGITDLLLV 147
+NRG IV+ L++ R++ ITD++++
Sbjct 115 LNRGNIVVGSLVEAARANDITDIVIL 140
> ECU11g1910
Length=269
Score = 56.2 bits (134), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 45/148 (30%), Positives = 72/148 (48%), Gaps = 29/148 (19%)
Query 5 LRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQV 64
+ R R R+EYL +K E +++ E+K+++ E LE+ +P +R
Sbjct 1 MNRRKRERREYLLQKEREMLDKEAEEKKRRLGEALESNTKIPHEIRK------------- 47
Query 65 LDLDDPQTLAPKTHVDDEYAYAG-VED----PSIALTTSRSPSSRLQQFVKELNLLLPNC 119
D P L DE Y G +E+ P + +TTSRSPSS+L F K L+L+L N
Sbjct 48 ---DAPMLL-------DEIIYEGRIEEEHRLPKVMVTTSRSPSSQLLHFAKHLSLVL-NG 96
Query 120 RRINRGTIVLDELLQLCRSSGITDLLLV 147
RG + +E+ + G T +++V
Sbjct 97 ENFIRGQLREEEVSDVAHKYGYTCVIIV 124
> At4g01560
Length=343
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 49/118 (41%), Gaps = 24/118 (20%)
Query 54 AAAAAARLAQVLDLDDPQTLAPKT------------HVDDEYAYAGVED----------- 90
A AA + A L + PQ + PKT DDE +A ++
Sbjct 66 ARDAAEKRALELGEEPPQKMIPKTIENTRESDETVCRPDDEELFADIDADEFNPVLRREI 125
Query 91 -PSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRSSGITDLLLV 147
P + LTT R S+R + EL ++PN RGT L ++++ T L++V
Sbjct 126 APKVLLTTCRFNSTRGPALISELLSVIPNSHYQKRGTYDLKKIVEYATKKDFTSLIVV 183
> Hs13376605
Length=349
Score = 37.0 bits (84), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Query 81 DEYA--YAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCRS 138
DE+A + P I +TTS P R + ++L+ ++PN R + L +++ C +
Sbjct 130 DEFASYFNKQTSPKILITTSDRPHGRTVRLCEQLSTVIPNSHVYYRRGLALKKIIPQCIA 189
Query 139 SGITDLLLV 147
TDL+++
Sbjct 190 RDFTDLIVI 198
> At2g11230
Length=962
Score = 35.0 bits (79), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 46/93 (49%), Gaps = 9/93 (9%)
Query 58 AARLAQVLDLDDPQTLAPKTHVDDEYAYAGVEDPS----IALTTSRSPS-SRLQQF---V 109
AA++ +++ D + P TH ++Y + ++D S AL +S S ++ +QF V
Sbjct 808 AAKILELVHGDLCGPITPSTHAGNKYIFVIIDDKSRYMWTALLKEKSDSFNKFKQFRALV 867
Query 110 KELNLLLPNCRRINRGT-IVLDELLQLCRSSGI 141
++ + R NRG +L E + C SGI
Sbjct 868 EQETRIKIQTFRTNRGGEFILHEFTKFCEESGI 900
> SPAC4F8.04
Length=306
Score = 33.9 bits (76), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 35/72 (48%), Gaps = 4/72 (5%)
Query 80 DDEYAYAGVED---PSIALTTSRSPSSRLQQFVKELNLLLPNCR-RINRGTIVLDELLQL 135
DDE++ E+ P + +TTS+ S + F EL PN R G I + E+ +
Sbjct 80 DDEFSAYFSEERKVPKLLVTTSKRASRKCYDFASELLDCFPNAEFRKRTGDIEVHEIAEA 139
Query 136 CRSSGITDLLLV 147
G TDLL++
Sbjct 140 AAKRGYTDLLVL 151
> CE28332
Length=384
Score = 30.8 bits (68), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 17/70 (24%), Positives = 35/70 (50%), Gaps = 2/70 (2%)
Query 80 DDEYAYAGVEDPSIALTTSRSPSSRLQ--QFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
+DE+A + S + + +P +++ +F EL +PN R ++L +++ +
Sbjct 164 NDEFAPYFNRETSPKVMITMTPKAKITTFKFCFELQKCIPNSEIFTRKNVLLKTIIEQAK 223
Query 138 SSGITDLLLV 147
TDLL+V
Sbjct 224 EREFTDLLVV 233
> Hs22054703
Length=355
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 26/94 (27%), Positives = 39/94 (41%), Gaps = 8/94 (8%)
Query 43 KPLPTNLRAAAAAAAAARLAQVLDLDDPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPS 102
P+P N R + L L+L+ TL T EY V+DP+ T RS
Sbjct 155 NPVPVNCRTGWFPVESQDL---LNLEGITTLLSTTEFSKEYKGRSVQDPACGTTGHRS-- 209
Query 103 SRLQQFVKELNLLLPNCRRINRGTIVLDELLQLC 136
L Q + L N +I++G ++ L+ C
Sbjct 210 --LPQLLCIERSLWDNVIKIDKG-LLFASALRFC 240
> At3g20150_2
Length=672
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 21/93 (22%), Positives = 45/93 (48%), Gaps = 6/93 (6%)
Query 43 KPLPTNLRAAAAAAAAARLAQVLDLDDPQTLAPKTHVD-DEYAYAGVEDPSIALTTSRSP 101
K P N R + ++ + +DD + + + V+ +++ + E S A S
Sbjct 98 KKFPVN-RDSVNSSFVTAFGESELMDDDEICSEEVEVEENDFGESLEEHDSAATVCKSSE 156
Query 102 SSRLQQFVKELNLLLPNCRRINRGTIVLDELLQ 134
SR+++FV E ++ + CR+ +++L E +Q
Sbjct 157 KSRIEEFVSENSISISPCRQ----SLILQEPIQ 185
> 7291031
Length=738
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 18/72 (25%), Positives = 37/72 (51%), Gaps = 3/72 (4%)
Query 9 ARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLD 68
A++ LFR++ ++ + +++ LE PT+L++++ AA A ++ D
Sbjct 152 AKVSDSVLFRQTRAAQDFSAEGPDESMEDPLEQE---PTDLKSSSPAAVAGKIELPKDEG 208
Query 69 DPQTLAPKTHVD 80
P+ AP +VD
Sbjct 209 APKVDAPAQNVD 220
> Hs16507950
Length=430
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 10 RLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLR 50
R+ E+ FRK L E E+ +E K Q K TLE L ++R
Sbjct 258 RVATEFAFRKRLREMEKVYSELKWQEKNTLEEIAELQEDIR 298
> CE05655
Length=430
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 31/120 (25%), Positives = 56/120 (46%), Gaps = 13/120 (10%)
Query 25 ERQLAERKQQVKETL-----ETGKPLPT--NLRAAAAAAAAARLAQVLDLDDPQTLAPKT 77
ER+LAE Q+ ET+ + K + T + A + + + ++ ++ + +T+A K+
Sbjct 205 ERRLAENHQKTHETITQAFDDMSKLMETAREMVALSKSISEKVRSRKGEISEDETIAFKS 264
Query 78 HVDDEYAYAGVEDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRGTIVLDELLQLCR 137
++ GV DP T S S Q+ KE++ +L + N G L E+ CR
Sbjct 265 YL----LSLGVSDPVTKSTFVGSDSEYFQKLAKEISDVLYEHIKENGGMCALPEV--YCR 318
> YDR464w
Length=1435
Score = 28.9 bits (63), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 31/45 (68%), Gaps = 4/45 (8%)
Query 6 RRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLR 50
R+TAR +E +K LE+ ++LAE ++++K+ +E G P P +LR
Sbjct 622 RQTAR--EERRHKKKLEK--QRLAEEQEELKKIVERGPPYPPDLR 662
> 7290873
Length=651
Score = 28.5 bits (62), Expect = 4.2, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 36/96 (37%), Gaps = 24/96 (25%)
Query 9 ARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAARLAQVLDLD 68
AR + L RK+L+ E + ++QQ+ TLE LD D
Sbjct 523 ARYDEVRLKRKTLQMAEEERVRKQQQLAPTLE------------------------LDPD 558
Query 69 DPQTLAPKTHVDDEYAYAGVEDPSIALTTSRSPSSR 104
+P D+E+ A E I L+ R S R
Sbjct 559 NPDQFVMMIEGDEEFRTAITEFQDIILSVERKRSKR 594
> YHR088w
Length=295
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 1/60 (1%)
Query 89 EDPSIALTTSRSPSSRLQQFVKELNLLLPNCRRINRG-TIVLDELLQLCRSSGITDLLLV 147
E P I LTT+ + +F L +LPN + R L E+ +C TD++++
Sbjct 91 EPPKIFLTTNVNAKKSAYEFANILIEILPNVTFVKRKFGYKLKEISDICIKRNFTDIVII 150
> SPBC12D12.03
Length=556
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 20 SLEEKERQLAERKQQVKETLETGKPLP 46
SL+E ER + + VK TLE+GK +P
Sbjct 389 SLDEMERSMHDSLSVVKRTLESGKVVP 415
> SPBC4B4.03
Length=803
Score = 28.5 bits (62), Expect = 5.1, Method: Composition-based stats.
Identities = 29/127 (22%), Positives = 48/127 (37%), Gaps = 19/127 (14%)
Query 1 GLKMLRRTARLRKEYLFRKSLEEKERQLAERKQQVKETLETGKPLPTNLRAAAAAAAAAR 60
G M+ R R + E L R+ L+ + L+ + + K + +R +
Sbjct 181 GRSMVGRDGRYKSEDLKRRKLQPSSKPLSSLEARAKVIMR-------QVRRYRDGSGRQL 233
Query 61 LAQVLDLDDPQTLAPKTHVDDEYAYAGVEDPS----IALTTSRSPSSRLQQFVKELNLLL 116
A L DP+ EY Y +E P I S+ ++QFV + NL+
Sbjct 234 FAPFERLPDPRMFP-------EY-YQAIEQPMALEVIQKKLSKHRYETIEQFVDDFNLMF 285
Query 117 PNCRRIN 123
N + N
Sbjct 286 DNAKSFN 292
> CE17349
Length=432
Score = 28.1 bits (61), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 2/35 (5%)
Query 19 KSLEEKERQLAERKQQVKETLET--GKPLPTNLRA 51
KSL E RQL + + +ET GKP+P + A
Sbjct 35 KSLTEMHRQLLTKTNMIDSVIETYNGKPIPAAIEA 69
> YGL150c
Length=1489
Score = 28.1 bits (61), Expect = 6.9, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 16 LFRKSLEEKERQLAERKQQVKETLETGKPLPTNLR 50
L R ++EE+ R A++K+QV++ + GK N++
Sbjct 1423 LVRGTIEERMRDRAKQKEQVQQVVMEGKTQEKNIK 1457
Lambda K H
0.317 0.132 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1785281974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40