bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3967_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
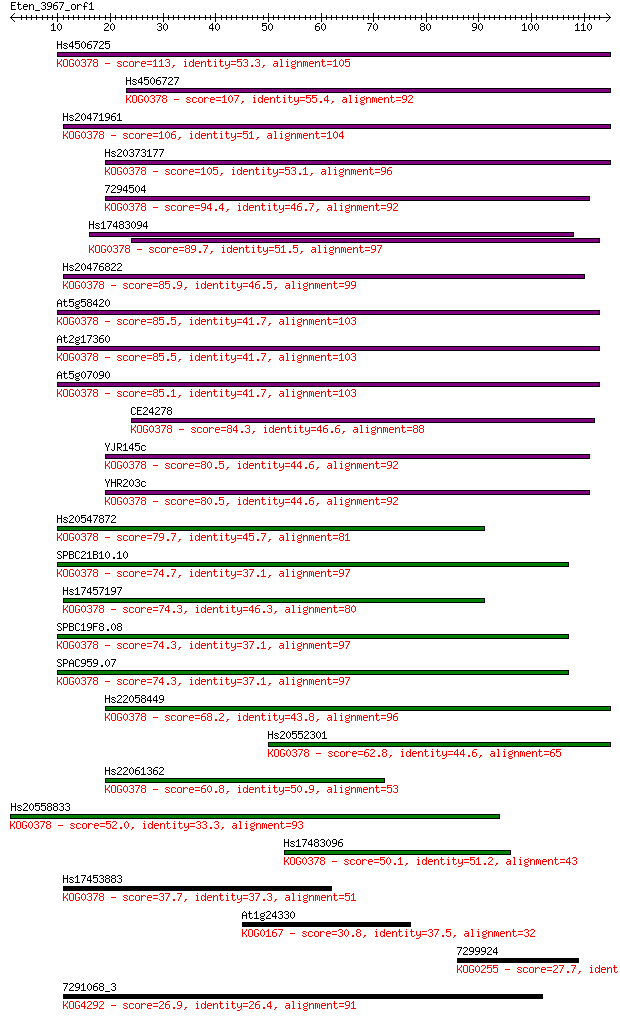
Hs4506725 113 9e-26
Hs4506727 107 6e-24
Hs20471961 106 1e-23
Hs20373177 105 2e-23
7294504 94.4 5e-20
Hs17483094 89.7 1e-18
Hs20476822 85.9 2e-17
At5g58420 85.5 2e-17
At2g17360 85.5 2e-17
At5g07090 85.1 3e-17
CE24278 84.3 5e-17
YJR145c 80.5 7e-16
YHR203c 80.5 7e-16
Hs20547872 79.7 1e-15
SPBC21B10.10 74.7 4e-14
Hs17457197 74.3 5e-14
SPBC19F8.08 74.3 6e-14
SPAC959.07 74.3 6e-14
Hs22058449 68.2 4e-12
Hs20552301 62.8 2e-10
Hs22061362 60.8 5e-10
Hs20558833 52.0 2e-07
Hs17483096 50.1 1e-06
Hs17453883 37.7 0.006
At1g24330 30.8 0.63
7299924 27.7 5.9
7291068_3 26.9 8.1
> Hs4506725
Length=263
Score = 113 bits (282), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 68/105 (64%), Gaps = 0/105 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD GN TGGAN G+IG I+N E PG FD+ H K ANGN+F
Sbjct 159 TIQIDLETGKITDFIKFDTGNLCMVTGGANLGRIGVITNRERHPGSFDVVHVKDANGNSF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
+ R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 219 ATRLSNIFVIGKGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> Hs4506727
Length=263
Score = 107 bits (267), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 51/92 (55%), Positives = 61/92 (66%), Gaps = 0/92 (0%)
Query 23 FFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKG 82
F KFD GN GGAN G++G I+N E PG FD+ H K ANGN+F+ R SNIF G G
Sbjct 172 FIKFDTGNLCMVIGGANLGRVGVITNRERHPGSFDVVHVKDANGNSFATRLSNIFVIGNG 231
Query 83 NKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
NKP + P GKGI L + E RDKRL +QS+G
Sbjct 232 NKPWISLPRGKGIRLTVAEERDKRLATKQSSG 263
> Hs20471961
Length=179
Score = 106 bits (265), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 53/104 (50%), Positives = 65/104 (62%), Gaps = 0/104 (0%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFS 70
++ + K + F KFD G TGGAN +IG I+N E PG FD+ K ANGN+F+
Sbjct 76 IKIDLETGKITDFIKFDAGKLCMVTGGANLERIGVITNRERHPGSFDVVRVKDANGNSFA 135
Query 71 PRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
R SNIF GKGNKP + P GKGI L I E RDKRL +QS+G
Sbjct 136 ARLSNIFVIGKGNKPWISLPRGKGIHLTIAEERDKRLAAKQSSG 179
> Hs20373177
Length=263
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 51/96 (53%), Positives = 61/96 (63%), Gaps = 0/96 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD GN GAN G++G I+N E PG D+ H K ANGN+F+ R SNIF
Sbjct 168 KITSFIKFDTGNVCMVIAGANLGRVGVITNRERHPGSCDVVHVKDANGNSFATRISNIFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
G GNKP + P GKGI L I E RDKRL +QS+G
Sbjct 228 IGNGNKPWISLPRGKGIRLTIAEERDKRLAAKQSSG 263
> 7294504
Length=261
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 43/92 (46%), Positives = 57/92 (61%), Gaps = 0/92 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + + KFD GN TGG N G++G + N E PG FD+ H K + G+ F+ R +N+F
Sbjct 168 KITDYIKFDSGNLCMITGGRNLGRVGTVVNRERHPGSFDIVHIKDSQGHVFATRLTNVFI 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQ 110
GKGNKP + P GKG+ L I E RDKRL +
Sbjct 228 IGKGNKPYISLPKGKGVKLSIAEERDKRLAAK 259
> Hs17483094
Length=608
Score = 89.7 bits (221), Expect = 1e-18, Method: Composition-based stats.
Identities = 49/92 (53%), Positives = 56/92 (60%), Gaps = 0/92 (0%)
Query 16 KLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSN 75
K K + F KFD GN G N G+IG I+N + G FD+ H K ANGN F+P SN
Sbjct 501 KTGKITDFIKFDTGNLCMVIGDTNLGRIGVITNRKKHRGSFDVVHVKDANGNRFAPWLSN 560
Query 76 IFFFGKGNKPGVFFPPGKGISLIIPEGRDKRL 107
IF GK NKP + P GKGI L I EGRDKRL
Sbjct 561 IFVTGKCNKPWISLPRGKGIRLTIAEGRDKRL 592
Score = 85.5 bits (210), Expect = 2e-17, Method: Composition-based stats.
Identities = 48/89 (53%), Positives = 54/89 (60%), Gaps = 4/89 (4%)
Query 24 FKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKGN 83
KFD GN TGGAN G+IG I+N E PG FD H K AN N+FS IF KGN
Sbjct 182 LKFDTGNLCLVTGGANWGRIGVITNRERHPGSFDTIHVKDANSNSFSI----IFVIDKGN 237
Query 84 KPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
KP + P GKGI I EGRDKRL P ++
Sbjct 238 KPCISLPRGKGIRFTIAEGRDKRLQPNRA 266
> Hs20476822
Length=183
Score = 85.9 bits (211), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 46/99 (46%), Positives = 56/99 (56%), Gaps = 5/99 (5%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFS 70
++ KS+ KFD GN + TGGAN G+IG I+N E PG FD+ H K AN N+F+
Sbjct 90 IQIDLDTGKSTDVIKFDTGNLWMVTGGANLGRIGVITNRERHPGSFDVVHVKDANSNSFA 149
Query 71 PRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPP 109
S+IF GKGNK GKGI L E DKRL
Sbjct 150 TWLSSIFVIGKGNK-----TQGKGIHLTTAEETDKRLAA 183
> At5g58420
Length=262
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 159 TIKLDLEANKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 219 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLASQQA 261
> At2g17360
Length=243
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 141 TIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 200
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 201 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLSAQQA 243
> At5g07090
Length=263
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 43/103 (41%), Positives = 55/103 (53%), Gaps = 0/103 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD GN TGG N G++G I N E G F+ H + + G+ F
Sbjct 160 TIKLDLEENKIVEFIKFDVGNVVMVTGGRNRGRVGVIKNREKHKGSFETIHIQDSTGHEF 219
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQS 112
+ R N++ GKG KP V P GKGI L I E KRL QQ+
Sbjct 220 ATRLGNVYTIGKGTKPWVSLPKGKGIKLTIIEEARKRLASQQA 262
> CE24278
Length=259
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 41/88 (46%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 24 FKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKGN 83
KF+PGN TGG N G++G I + E PG D+ H K + G++F+ R SN+F GKGN
Sbjct 172 VKFEPGNLAYVTGGRNVGRVGIIGHRERLPGASDIIHIKDSAGHSFATRISNVFVIGKGN 231
Query 84 KPGVFFPPGKGISLIIPEGRDKRLPPQQ 111
K V P G GI L I E RDKR+ +
Sbjct 232 KALVSLPTGAGIRLSIAEERDKRMAQKH 259
> YJR145c
Length=261
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD G TGG N G+IG I + E G FDL H K + N F R +N+F
Sbjct 168 KITDFIKFDAGKLVYVTGGRNLGRIGTIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQ 110
G+ KP + P GKGI L I E RD+R Q
Sbjct 228 IGEQGKPYISLPKGKGIKLSIAEERDRRRAQQ 259
> YHR203c
Length=261
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 0/92 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KFD G TGG N G+IG I + E G FDL H K + N F R +N+F
Sbjct 168 KITDFIKFDAGKLVYVTGGRNLGRIGTIVHKERHDGGFDLVHIKDSLDNTFVTRLNNVFV 227
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQ 110
G+ KP + P GKGI L I E RD+R Q
Sbjct 228 IGEQGKPYISLPKGKGIKLSIAEERDRRRAQQ 259
> Hs20547872
Length=153
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 50/81 (61%), Gaps = 0/81 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
++ + K + F KFD G+ TGGAN G+IG I+N E PG FD+ H + ANGN+F
Sbjct 73 TIQIDLETGKITDFIKFDTGHLCMVTGGANLGRIGVITNRERHPGSFDIVHMEDANGNSF 132
Query 70 SPRFSNIFFFGKGNKPGVFFP 90
+ + SNIF GKGNK + P
Sbjct 133 ATQLSNIFVIGKGNKLWISLP 153
> SPBC21B10.10
Length=262
Score = 74.7 bits (182), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 49/97 (50%), Gaps = 0/97 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD TGG N G++G I + E G F++ H K A F
Sbjct 159 TIKLNLETNKIESFIKFDTSAQVMVTGGRNMGRVGTIVHREHHLGSFEIIHVKDALDREF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKR 106
+ R SN+F G+ K + P GKG+ L I E RD+R
Sbjct 219 ATRLSNVFVIGEAGKSWISLPKGKGVKLSITEERDRR 255
> Hs17457197
Length=167
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 37/80 (46%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFS 70
+ ++ K + F KFD GN T GAN G+ G I+N E G FD+ H K ANGN+F+
Sbjct 88 IQTDLEIGKITNFIKFDNGNLCMVTVGANLGRTGVITNRERHSGSFDVVHMKDANGNSFA 147
Query 71 PRFSNIFFFGKGNKPGVFFP 90
+ SNIF GKGNKP P
Sbjct 148 TQLSNIFVIGKGNKPQTSLP 167
> SPBC19F8.08
Length=262
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 49/97 (50%), Gaps = 0/97 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD TGG N G++G I + E G F++ H K A F
Sbjct 159 TIKLNLETNKIESFIKFDTSAQVMVTGGRNMGRVGTIVHREHHLGSFEIIHVKDALDREF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKR 106
+ R SN+F G+ K + P GKG+ L I E RD+R
Sbjct 219 ATRLSNVFVIGETGKSWISLPKGKGVKLSITEERDRR 255
> SPAC959.07
Length=262
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/97 (37%), Positives = 49/97 (50%), Gaps = 0/97 (0%)
Query 10 PFRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNF 69
+L + K F KFD TGG N G++G I + E G F++ H K A F
Sbjct 159 TIKLNLETNKIESFIKFDTSAQVMVTGGRNMGRVGTIVHREHHLGSFEIIHVKDALDREF 218
Query 70 SPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKR 106
+ R SN+F G+ K + P GKG+ L I E RD+R
Sbjct 219 ATRLSNVFVIGETGKSWISLPKGKGVKLSITEERDRR 255
> Hs22058449
Length=160
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 42/96 (43%), Positives = 52/96 (54%), Gaps = 14/96 (14%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSPRFSNIFF 78
K + F KF+ N TGGAN G+IG I+N E PG FD+ H K AN SNI
Sbjct 79 KITGFIKFNTENLCMVTGGANLGRIGVITNRERHPGSFDVVHGKDAN--------SNI-- 128
Query 79 FGKGNKPGVFFPPGKGISLIIPEGRDKRLPPQQSNG 114
NKP + P GK + L I E DKRL +Q++G
Sbjct 129 ----NKPWISLPQGKVMRLTIAEETDKRLVAKQTSG 160
> Hs20552301
Length=297
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 29/65 (44%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 50 EGPPGFFDLAHWKKANGNNFSPRFSNIFFFGKGNKPGVFFPPGKGISLIIPEGRDKRLPP 109
E G F++ H K NGN+F+ SNIF KGNKP + P GKGI I E +D+R+
Sbjct 233 ERHSGCFNVVHVKHTNGNSFATWLSNIFVISKGNKPWISLPQGKGIHFTIAEEKDERMMA 292
Query 110 QQSNG 114
+ S+G
Sbjct 293 KPSSG 297
> Hs22061362
Length=173
Score = 60.8 bits (146), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 27/53 (50%), Positives = 35/53 (66%), Gaps = 0/53 (0%)
Query 19 KSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHWKKANGNNFSP 71
K + F +F+ GN TGGAN G+IG I+N E PGFFD+ H K AN N+F+
Sbjct 121 KITDFIRFNTGNLCMVTGGANLGRIGAITNREKHPGFFDVVHVKDANSNSFAT 173
> Hs20558833
Length=109
Score = 52.0 bits (123), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 47/97 (48%), Gaps = 4/97 (4%)
Query 1 LIPSSRGMIPFRLPW--KLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNP--EGPPGFF 56
++ + R + P + W + + S F K F AN +IG S E P FF
Sbjct 1 MLKTDRCVCPLDIHWPYQASPSHRFPKKQAQVCFLQRRNANLEQIGKESTRQRERQPDFF 60
Query 57 DLAHWKKANGNNFSPRFSNIFFFGKGNKPGVFFPPGK 93
++ H K NGN+F+ + SNIF G+ N+P + P K
Sbjct 61 NMVHVKDVNGNSFAIQLSNIFIIGEDNRPWIPCPKEK 97
> Hs17483096
Length=371
Score = 50.1 bits (118), Expect = 1e-06, Method: Composition-based stats.
Identities = 22/43 (51%), Positives = 30/43 (69%), Gaps = 0/43 (0%)
Query 53 PGFFDLAHWKKANGNNFSPRFSNIFFFGKGNKPGVFFPPGKGI 95
PG FD+ H K AN N+F+ + SN+F GKG+KP + P GKG+
Sbjct 323 PGCFDVVHVKDANSNSFATQLSNLFVIGKGSKPWISLPRGKGV 365
> Hs17453883
Length=105
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 25/51 (49%), Gaps = 3/51 (5%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAHW 61
++ + K + F KFD GN TG AN G+IG I+ G DL W
Sbjct 56 IQIDLETGKITDFTKFDTGNLCMVTGSANLGRIGVITTERGT---VDLLTW 103
> At1g24330
Length=709
Score = 30.8 bits (68), Expect = 0.63, Method: Composition-based stats.
Identities = 12/32 (37%), Positives = 19/32 (59%), Gaps = 0/32 (0%)
Query 45 GISNPEGPPGFFDLAHWKKANGNNFSPRFSNI 76
GI+ P GPP DL +W+ A ++ SP ++
Sbjct 284 GITVPTGPPESLDLNYWRLAMSDSESPNSKSV 315
> 7299924
Length=556
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 10/23 (43%), Positives = 16/23 (69%), Gaps = 0/23 (0%)
Query 86 GVFFPPGKGISLIIPEGRDKRLP 108
G+ F G + L++PE R+K+LP
Sbjct 485 GLLFLSGAYVCLLLPETRNKKLP 507
> 7291068_3
Length=3710
Score = 26.9 bits (58), Expect = 8.1, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 36/93 (38%), Gaps = 39/93 (41%)
Query 11 FRLPWKLAKSSYFFKFDPGNPFKGTGGANPGKIGGISNPEGPPGFFDLAH--WKKANGNN 68
FRL W++ F F G GG+ +G IS+P P + ++AH W+
Sbjct 1199 FRLRWRI------FAF-------GCGGSLRSNMGAISSPRYPNSYPNMAHCEWR------ 1239
Query 69 FSPRFSNIFFFGKGNKPGVFFPPGKGISLIIPE 101
+ PG GISL+I +
Sbjct 1240 ------------------ISLHPGSGISLLIED 1254
Lambda K H
0.320 0.146 0.483
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40