bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_3968_orf3
Length=159
Score E
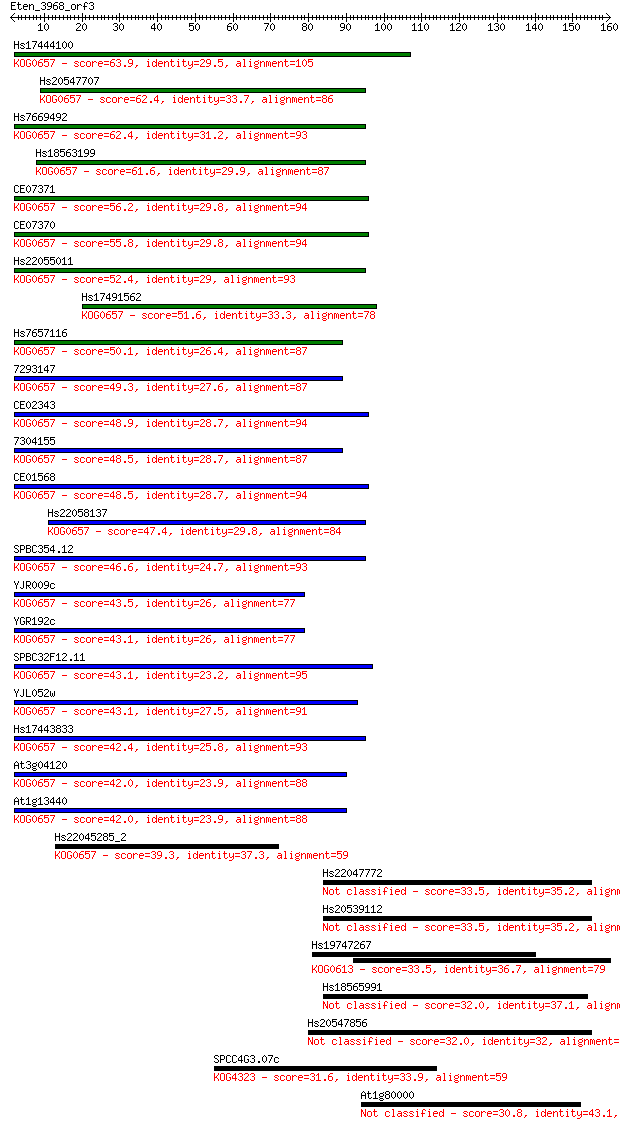
Sequences producing significant alignments: (Bits) Value
Hs17444100 63.9 1e-10
Hs20547707 62.4 3e-10
Hs7669492 62.4 3e-10
Hs18563199 61.6 6e-10
CE07371 56.2 3e-08
CE07370 55.8 3e-08
Hs22055011 52.4 4e-07
Hs17491562 51.6 7e-07
Hs7657116 50.1 2e-06
7293147 49.3 3e-06
CE02343 48.9 4e-06
7304155 48.5 5e-06
CE01568 48.5 6e-06
Hs22058137 47.4 1e-05
SPBC354.12 46.6 2e-05
YJR009c 43.5 2e-04
YGR192c 43.1 2e-04
SPBC32F12.11 43.1 2e-04
YJL052w 43.1 3e-04
Hs17443833 42.4 4e-04
At3g04120 42.0 5e-04
At1g13440 42.0 6e-04
Hs22045285_2 39.3 0.003
Hs22047772 33.5 0.17
Hs20539112 33.5 0.17
Hs19747267 33.5 0.19
Hs18565991 32.0 0.54
Hs20547856 32.0 0.55
SPCC4G3.07c 31.6 0.62
At1g80000 30.8 1.1
> Hs17444100
Length=324
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 31/105 (29%), Positives = 48/105 (45%), Gaps = 0/105 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP ++ +I+KVVK +GP KG+ + Q F T +F GI
Sbjct 220 VINLTCHLEKPAKYDDIKKVVKQTSEGPLKGILGYTEHQAVSSNFNSNTHSSTFNVRAGI 279
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKGKGSRAPPPPRG 106
+L F K+ W+ +F ++ L+ A K K +R P P R
Sbjct 280 ALNNHFVKIISWYSDEFGYSNKVVDLMAHMASKRKTTRPPAPERA 324
> Hs20547707
Length=303
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/86 (33%), Positives = 44/86 (51%), Gaps = 0/86 (0%)
Query 9 LGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGISLKGPFF 68
L KP ++ +I+KVVK +GP KG+ + G QV F T +F G G +L F
Sbjct 217 LEKPAKYDDIKKVVKQASEGPLKGILGYTGHQVVSSDFNSDTHSSTFNAGAGTALNDHFV 276
Query 69 KVFFWFEKKFCSPHRGGGLLVPKAPK 94
K+ W++ +F +R L+ A K
Sbjct 277 KLISWYDNEFGYNNRVVDLMAHMASK 302
> Hs7669492
Length=335
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 29/93 (31%), Positives = 45/93 (48%), Gaps = 0/93 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP ++ +I+KVVK +GP KG+ + QV F T +F G GI
Sbjct 242 VVDLTCRLEKPAKYDDIKKVVKQASEGPLKGILGYTEHQVVSSDFNSDTHSSTFDAGAGI 301
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPK 94
+L F K+ W++ +F +R L+ A K
Sbjct 302 ALNDHFVKLISWYDNEFGYSNRVVDLMAHMASK 334
> Hs18563199
Length=243
Score = 61.6 bits (148), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 45/87 (51%), Gaps = 0/87 (0%)
Query 8 PLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGISLKGPF 67
P GKP ++++I+K++K + P KG+ + Q+ F T +F G GI+L F
Sbjct 156 PSGKPAKYYDIKKMMKQASEDPIKGILGYTEHQIVSSDFNSDTQSSTFDAGAGITLNDHF 215
Query 68 FKVFFWFEKKFCSPHRGGGLLVPKAPK 94
K+ W++ +F +R L+ A K
Sbjct 216 VKLISWYDNEFGYSNRVVDLMAHMASK 242
> CE07371
Length=341
Score = 56.2 bits (134), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 40/94 (42%), Gaps = 0/94 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP +I+KV+K GP KG+ + QV F T F G I
Sbjct 248 VVDLTARLEKPASLDDIKKVIKAAADGPMKGILAYTEDQVVSTDFVSDTNSSIFDAGASI 307
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKG 95
SL F K+ W++ +F +R L+ A K
Sbjct 308 SLNPHFVKLVSWYDNEFGYSNRVVDLISYIATKA 341
> CE07370
Length=341
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 40/94 (42%), Gaps = 0/94 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP +I+KV+K GP KG+ + QV F T F G I
Sbjct 248 VVDLTARLEKPASLDDIKKVIKAAADGPMKGILAYTEDQVVSTDFVSDTNSSIFDAGASI 307
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKG 95
SL F K+ W++ +F +R L+ A K
Sbjct 308 SLNPHFVKLVSWYDNEFGYSNRVVDLISYIATKA 341
> Hs22055011
Length=288
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 43/95 (45%), Gaps = 2/95 (2%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGT--PFFSFGGGG 59
+ LGK ++ I+KVV +GP KG+ + QV F T F+ G
Sbjct 193 VVDLTCCLGKTAKYDGIEKVVGQASEGPLKGILGYTEHQVVSSEFNSNTHSSAFNVNEGN 252
Query 60 GISLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPK 94
I+L F K+ W++ +F + +R L+V K
Sbjct 253 DIALNNHFVKLISWYDNEFGNSNRVIDLMVHLTSK 287
> Hs17491562
Length=242
Score = 51.6 bits (122), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 26/78 (33%), Positives = 37/78 (47%), Gaps = 0/78 (0%)
Query 20 KVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGISLKGPFFKVFFWFEKKFC 79
KVVK +G +G+ + QV F T +F G GI+L F K+ W+E +F
Sbjct 165 KVVKQASEGTLEGILGYTEHQVVASDFNSITHSSTFKAGVGIALNNHFVKLISWYENEFG 224
Query 80 SPHRGGGLLVPKAPKGKG 97
+R L+V A KG
Sbjct 225 YSNRVVDLMVHMASKGSS 242
> Hs7657116
Length=408
Score = 50.1 bits (118), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 23/87 (26%), Positives = 37/87 (42%), Gaps = 0/87 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L +P + I++ VK KGP G+ + +V F T F GI
Sbjct 314 VVDLTCRLAQPAPYSAIKEAVKAAAKGPMAGILAYTEDEVVSTDFLGDTHSSIFDAKAGI 373
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLL 88
+L F K+ W++ ++ HR LL
Sbjct 374 ALNDNFVKLISWYDNEYGYSHRVVDLL 400
> 7293147
Length=332
Score = 49.3 bits (116), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 37/87 (42%), Gaps = 0/87 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ LGK + EI+ V+ GP KG+ + +V F T F GI
Sbjct 239 VVDLTVRLGKGASYDEIKAKVQEAANGPLKGILGYTDEEVVSTDFLSDTHSSVFDAKAGI 298
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLL 88
SL F K+ W++ +F +R L+
Sbjct 299 SLNDKFVKLISWYDNEFGYSNRVIDLI 325
> CE02343
Length=341
Score = 48.9 bits (115), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 40/94 (42%), Gaps = 0/94 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP +I+KVVK GP KG+ + QV F F G I
Sbjct 248 VVDLTVRLEKPASMDDIKKVVKAAADGPMKGILAYTEDQVVSTDFVSDPHSSIFDAGACI 307
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKG 95
SL F K+ W++ ++ +R L+ A +G
Sbjct 308 SLNPNFVKLVSWYDNEYGYSNRVVDLIGYIATRG 341
> 7304155
Length=331
Score = 48.5 bits (114), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 38/87 (43%), Gaps = 0/87 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ LGK + EI+ V+ KGP KG+ + +V F T F GI
Sbjct 238 VVDLTVRLGKGATYDEIKAKVEEASKGPLKGILGYTDEEVVSTDFFSDTHSSVFDAKAGI 297
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLL 88
SL F K+ W++ +F +R L+
Sbjct 298 SLNDKFVKLISWYDNEFGYSNRVIDLI 324
> CE01568
Length=341
Score = 48.5 bits (114), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 27/94 (28%), Positives = 40/94 (42%), Gaps = 0/94 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP +I+KVVK GP KG+ + QV F F G I
Sbjct 248 VVDLTVRLEKPASMDDIKKVVKAAADGPMKGILAYTEDQVVSTDFVSDPHSSIFDTGACI 307
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKG 95
SL F K+ W++ ++ +R L+ A +G
Sbjct 308 SLNPNFVKLVSWYDNEYGYSNRVVDLIGYIATRG 341
> Hs22058137
Length=307
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query 11 KPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGISLKGPFFKV 70
KP ++ +I+KV +GP KG+ + QV F T + F G G++L F K+
Sbjct 228 KPGKYDDIKKV-----EGPLKGILGYNEHQVVSFDFNSNTQYSIFDAGVGVTLNDHFVKL 282
Query 71 FFWFEKKFCSPHRGGGLLVPKAPK 94
++ +F +R L+V A K
Sbjct 283 ISCYDNEFGCSNRVVDLMVHIASK 306
> SPBC354.12
Length=335
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 37/93 (39%), Gaps = 0/93 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP + +I+ +K +GP KG+ + V F F GI
Sbjct 242 VVDLTVKLAKPTNYEDIKAAIKAASEGPMKGVLGYTEDSVVSTDFCGDNHSSIFDASAGI 301
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPK 94
L F K+ W++ ++ HR L+ A K
Sbjct 302 QLSPQFVKLVSWYDNEWGYSHRVVDLVAYTASK 334
> YJR009c
Length=332
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 32/77 (41%), Gaps = 0/77 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L K + EI+KVVK +G KG+ + V F + F GI
Sbjct 240 VVDLTVKLNKETTYDEIKKVVKAAAEGKLKGVLGYTEDAVVSSDFLGDSNSSIFDAAAGI 299
Query 62 SLKGPFFKVFFWFEKKF 78
L F K+ W++ ++
Sbjct 300 QLSPKFVKLVSWYDNEY 316
> YGR192c
Length=332
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 20/77 (25%), Positives = 32/77 (41%), Gaps = 0/77 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L K + EI+KVVK +G KG+ + V F + F GI
Sbjct 240 VVDLTVKLNKETTYDEIKKVVKAAAEGKLKGVLGYTEDAVVSSDFLGDSHSSIFDASAGI 299
Query 62 SLKGPFFKVFFWFEKKF 78
L F K+ W++ ++
Sbjct 300 QLSPKFVKLVSWYDNEY 316
> SPBC32F12.11
Length=336
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 36/95 (37%), Gaps = 0/95 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP + +I+ +K +GP KG+ + V F F GI
Sbjct 242 VVDLTVKLAKPTNYEDIKAAIKAASEGPMKGVLGYTEDAVVSTDFCGDNHSSIFDASAGI 301
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKGK 96
L F K+ W++ ++ R L+ A K
Sbjct 302 QLSPQFVKLVSWYDNEWGYSRRVVDLVAYTAAKDN 336
> YJL052w
Length=332
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 38/93 (40%), Gaps = 2/93 (2%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L K + +I+K VK +GP KG+ + V F T F GI
Sbjct 240 VVDLTVKLEKEATYDQIKKAVKAAAEGPMKGVLGYTEDAVVSSDFLGDTHASIFDASAGI 299
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLL--VPKA 92
L F K+ W++ ++ R L+ V KA
Sbjct 300 QLSPKFVKLISWYDNEYGYSARVVDLIEYVAKA 332
> Hs17443833
Length=268
Score = 42.4 bits (98), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 40/93 (43%), Gaps = 11/93 (11%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L KP ++ +I+K G++ QV F + T +F G GI
Sbjct 186 VMDLTCRLEKPAKYDDIKK-----------GIWGDTEHQVVSSDFNRDTHSSTFDAGAGI 234
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPK 94
+LK F K+ W++ +F +R L+ A K
Sbjct 235 ALKDHFVKLISWYDNEFGYSNRVVDLMAHIASK 267
> At3g04120
Length=338
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 35/88 (39%), Gaps = 0/88 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L K + EI+K +K +G KG+ + V F F GI
Sbjct 246 VVDLTVRLEKAATYDEIKKAIKEESEGKLKGILGYTEDDVVSTDFVGDNRSSIFDAKAGI 305
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLV 89
+L F K+ W++ ++ R L+V
Sbjct 306 ALSDKFVKLVSWYDNEWGYSSRVVDLIV 333
> At1g13440
Length=338
Score = 42.0 bits (97), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 35/88 (39%), Gaps = 0/88 (0%)
Query 2 FLFFPPPLGKPPQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFSFGGGGGI 61
+ L K + EI+K +K +G KG+ + V F F GI
Sbjct 246 VVDLTVRLEKAATYDEIKKAIKEESEGKMKGILGYTEDDVVSTDFVGDNRSSIFDAKAGI 305
Query 62 SLKGPFFKVFFWFEKKFCSPHRGGGLLV 89
+L F K+ W++ ++ R L+V
Sbjct 306 ALSDKFVKLVSWYDNEWGYSSRVVDLIV 333
> Hs22045285_2
Length=246
Score = 39.3 bits (90), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 30/60 (50%), Gaps = 10/60 (16%)
Query 13 PQFFEIQKVVKPGLKGPFKGLFWFPGGQVFFLVFKKGTPFFS-FGGGGGISLKGPFFKVF 71
P++ +I+KVVK + KGL + Q TP FS F G GI+LK F K+
Sbjct 195 PKYDDIKKVVKLASEATLKGLLSYTEHQ---------TPTFSTFNAGNGIALKDHFVKLI 245
> Hs22047772
Length=777
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 33/85 (38%), Gaps = 20/85 (23%)
Query 84 GGGLLVPKAPKGKGSRAPPPPRGP---------EGKVSFLWGGSSPFSPPGKFPGFPNFP 134
G G + P +G P RGP ++G S + PP FPG P+ P
Sbjct 690 GPGFIPPPLAPVRGPLFPVDTRGPFMRRGPPFPPPPPGTMFGASRGYFPPRDFPGPPHAP 749
Query 135 F-----LKPRKRGGVLGPPPFFPPK 154
F PR G PP+F P+
Sbjct 750 FAMRNIYPPR------GLPPYFHPR 768
> Hs20539112
Length=777
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 33/85 (38%), Gaps = 20/85 (23%)
Query 84 GGGLLVPKAPKGKGSRAPPPPRGP---------EGKVSFLWGGSSPFSPPGKFPGFPNFP 134
G G + P +G P RGP ++G S + PP FPG P+ P
Sbjct 690 GPGFIPPPLAPVRGPLFPVDTRGPFMRRGPPFPPPPPGTMFGASRGYFPPRDFPGPPHAP 749
Query 135 F-----LKPRKRGGVLGPPPFFPPK 154
F PR G PP+F P+
Sbjct 750 FAMRNIYPPR------GLPPYFHPR 768
> Hs19747267
Length=34350
Score = 33.5 bits (75), Expect = 0.19, Method: Composition-based stats.
Identities = 20/61 (32%), Positives = 29/61 (47%), Gaps = 5/61 (8%)
Query 81 PHRGGGLLVPKAPKGKGSRAPPPPRG--PEGKVSFLWGGSSPFSPPGKFPGFPNFPFLKP 138
P + L+VPK P+ ++ P P+ PE KV+ P PP K P P P L+
Sbjct 11657 PEKKVPLVVPKKPEAPPAKVPEVPKEVVPEKKVAV---PKKPEVPPAKVPEVPKKPVLEE 11713
Query 139 R 139
+
Sbjct 11714 K 11714
Score = 28.1 bits (61), Expect = 7.0, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 30/75 (40%), Gaps = 12/75 (16%)
Query 92 APKGKGSRAPPPPRGPEGKVSFLWGGSSPFSPPGKFPGFPNF-------PFLKPRKRGGV 144
AP K P P PE KV + +PP K P P P P+K V
Sbjct 10954 APPAKVPEVPKKPV-PEKKVP-VPAPKKVEAPPAKVPEVPKKLIPEEKKPTPVPKK---V 11008
Query 145 LGPPPFFPPKKIPLP 159
PPP P K+ P+P
Sbjct 11009 EAPPPKVPKKREPVP 11023
> Hs18565991
Length=777
Score = 32.0 bits (71), Expect = 0.54, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 31/84 (36%), Gaps = 14/84 (16%)
Query 84 GGGLLVPKAPKGKGSRAPPPPRGP---------EGKVSFLWGGSSPFSPPGKFPGFPNFP 134
G GL+ P G P RGP ++G S + PP FPG P+ P
Sbjct 690 GPGLIPPPLAPISGPLFPVDTRGPFMRRGPPFPPPPPGTMFGASRGYFPPRDFPGPPHAP 749
Query 135 F-----LKPRKRGGVLGPPPFFPP 153
F PR L P P F P
Sbjct 750 FAMRNIYPPRGLPPYLHPRPGFYP 773
> Hs20547856
Length=775
Score = 32.0 bits (71), Expect = 0.55, Method: Composition-based stats.
Identities = 24/84 (28%), Positives = 32/84 (38%), Gaps = 10/84 (11%)
Query 80 SPHRGGGLLVPKAPKGKGSRAPPPPRGP---------EGKVSFLWGGSSPFSPPGKFPGF 130
S G G + P +G P RGP ++G S + PP PG
Sbjct 683 SEATGPGFVPPPLAPIRGPLFPVDTRGPFMRRGPPFPPPPPGTMFGASPDYFPPRDVPGP 742
Query 131 PNFPFLKPRKRGGVLGPPPFFPPK 154
P PF R + G PP+ PP+
Sbjct 743 PRAPFAM-RNVCPLRGFPPYLPPR 765
> SPCC4G3.07c
Length=461
Score = 31.6 bits (70), Expect = 0.62, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 29/59 (49%), Gaps = 3/59 (5%)
Query 55 FGGGGGISLKGPFFKVFFWFEKKFCSPHRGGGLLVPKAPKGKGSRAPPPPRGPEGKVSF 113
G GIS GPF F ++ +P+ GG+ +P A G+ + PP + P VSF
Sbjct 53 VASGSGISDVGPFGADFHQLQQHVQTPY--GGMTMP-ASSSSGATSVPPEQDPSLSVSF 108
> At1g80000
Length=656
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 25/61 (40%), Positives = 30/61 (49%), Gaps = 9/61 (14%)
Query 94 KGKGSRAPPPPRGPEGKVSFLWGGSSPFSPPGKFPGF--PNFP-FLKPRKRGGVLGPPPF 150
KGKGS P G+ SF++GG+ P G G PNFP FL + GG G P
Sbjct 444 KGKGSLQP------SGRGSFMYGGTQFMGPAGMAAGHGNPNFPAFLPVMQFGGQHGGVPT 497
Query 151 F 151
F
Sbjct 498 F 498
Lambda K H
0.326 0.156 0.548
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40