bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4030_orf2
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
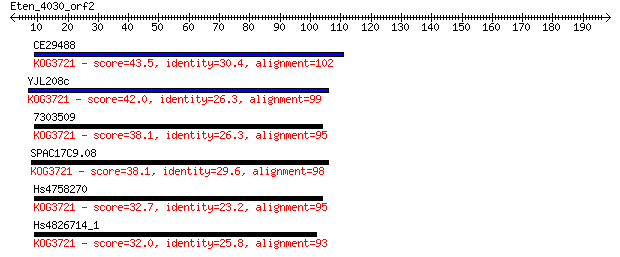
CE29488 43.5 3e-04
YJL208c 42.0 8e-04
7303509 38.1 0.011
SPAC17C9.08 38.1 0.012
Hs4758270 32.7 0.48
Hs4826714_1 32.0 0.81
> CE29488
Length=308
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 31/102 (30%), Positives = 44/102 (43%), Gaps = 31/102 (30%)
Query 9 IEYEVLGHGFLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYSTPPP 68
I+Y+V+G + THFFKV L TP
Sbjct 221 IKYQVIGDNNVAVPTHFFKVALF-------------------------------EVTPGK 249
Query 69 AVLGAFVLPNRVIYEDVRISRFEVPLSFVELVSGIDLGVVLD 110
L +++LPN VI + V IS+F VPL VE +G+++ LD
Sbjct 250 FELESYILPNAVIEDTVEISKFHVPLDAVERSAGLEIFARLD 291
> YJL208c
Length=329
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 47/100 (47%), Gaps = 27/100 (27%)
Query 7 MQIEYEVLGHG-FLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYST 65
++ YEV+G+ + THFFK+++A +P +E +
Sbjct 210 FRVNYEVIGNPPSIAVPTHFFKLIVAEAPTANPAREDIA--------------------- 248
Query 66 PPPAVLGAFVLPNRVIYEDVRISRFEVPLSFVELVSGIDL 105
+ AFVLPN I + +++ FEVP+ +E +G++L
Sbjct 249 -----VAAFVLPNEPISNETKLTDFEVPIDALERSTGLEL 283
> 7303509
Length=310
Score = 38.1 bits (87), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 40/95 (42%), Gaps = 31/95 (32%)
Query 9 IEYEVLGHGFLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYSTPPP 68
++YEV+G + THF+KV++ S + E
Sbjct 228 VKYEVIGANTVAVPTHFYKVIVGESADHKLHME--------------------------- 260
Query 69 AVLGAFVLPNRVIYEDVRISRFEVPLSFVELVSGI 103
++V+PN+VI D IS F+VP VE +G+
Sbjct 261 ----SYVMPNQVISNDTPISVFQVPPESVERSAGL 291
> SPAC17C9.08
Length=335
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 46/99 (46%), Gaps = 29/99 (29%)
Query 8 QIEYEVLGHG-FLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYSTP 66
+++Y V+G+ + THFFKV++A E P S+P
Sbjct 214 EVQYRVIGNPPNVAVPTHFFKVIIAEKSGE-------------------------PTSSP 248
Query 67 PPAVLGAFVLPNRVIYEDVRISRFEVPLSFVELVSGIDL 105
A AFVLPN+ I ++ + F VP+ VE SG+++
Sbjct 249 SVA---AFVLPNKPIADNFPLKNFAVPVEVVERASGLEI 284
> Hs4758270
Length=297
Score = 32.7 bits (73), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 22/95 (23%), Positives = 38/95 (40%), Gaps = 31/95 (32%)
Query 9 IEYEVLGHGFLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYSTPPP 68
++Y+V+G + THFFKVL+ + +
Sbjct 213 VKYQVIGKNHVAVPTHFFKVLILEAAGGQIE----------------------------- 243
Query 69 AVLGAFVLPNRVIYEDVRISRFEVPLSFVELVSGI 103
L +V+PN + E + + RF VP+ +E SG+
Sbjct 244 --LRTYVMPNAPVDEAIPLERFLVPIESIERASGL 276
> Hs4826714_1
Length=308
Score = 32.0 bits (71), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 40/93 (43%), Gaps = 28/93 (30%)
Query 9 IEYEVLGHGFLPAATHFFKVLLAVSPKEAVRKERLGASPSGSHKQSQAKKDYWPYSTPPP 68
+ Y+V+G + +H +KV+LA + +V E P
Sbjct 212 VSYQVIGEDNVAVPSHLYKVILA--RRSSVSTE--------------------------P 243
Query 69 AVLGAFVLPNRVIYEDVRISRFEVPLSFVELVS 101
LGAFV+PN I +++ F+V L +E +S
Sbjct 244 LALGAFVVPNEAIGFQPQLTEFQVSLQDLEKLS 276
Lambda K H
0.320 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3407623970
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40