bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
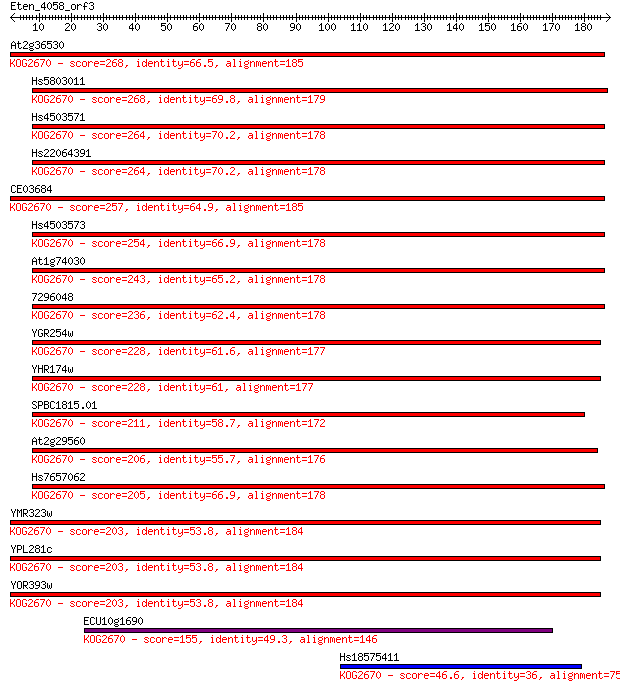
Query= Eten_4058_orf3
Length=187
Score E
Sequences producing significant alignments: (Bits) Value
At2g36530 268 4e-72
Hs5803011 268 7e-72
Hs4503571 264 8e-71
Hs22064391 264 8e-71
CE03684 257 1e-68
Hs4503573 254 5e-68
At1g74030 243 1e-64
7296048 236 1e-62
YGR254w 228 4e-60
YHR174w 228 5e-60
SPBC1815.01 211 7e-55
At2g29560 206 3e-53
Hs7657062 205 4e-53
YMR323w 203 2e-52
YPL281c 203 2e-52
YOR393w 203 2e-52
ECU10g1690 155 4e-38
Hs18575411 46.6 3e-05
> At2g36530
Length=444
Score = 268 bits (686), Expect = 4e-72, Method: Compositional matrix adjust.
Identities = 123/185 (66%), Positives = 149/185 (80%), Gaps = 0/185 (0%)
Query 1 YSKEAKSYDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALT 60
+ E K+YDL+FK + + ++GD LKDL+K + E+PIVSIEDPFDQDD+ YA +T
Sbjct 256 FYSEDKTYDLNFKEENNNGSQKISGDALKDLYKSFVAEYPIVSIEDPFDQDDWEHYAKMT 315
Query 61 AEIGSKVQVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWG 120
E G++VQ+VGDDLLVTNP R+ KA+ K+CNALLLKVNQIGS+TE+IEA K+++ +GWG
Sbjct 316 TECGTEVQIVGDDLLVTNPKRVAKAIAEKSCNALLLKVNQIGSVTESIEAVKMSKKAGWG 375
Query 121 VMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGE 180
VM SHRSGETED+FIADL VGL TGQIKTGAPCRSERL KYNQLLRIEE+L YAG
Sbjct 376 VMTSHRSGETEDTFIADLAVGLSTGQIKTGAPCRSERLAKYNQLLRIEEELGSEAIYAGV 435
Query 181 NFRNP 185
NFR P
Sbjct 436 NFRKP 440
> Hs5803011
Length=434
Score = 268 bits (684), Expect = 7e-72, Method: Compositional matrix adjust.
Identities = 125/179 (69%), Positives = 151/179 (84%), Gaps = 3/179 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFKSP D R +TGDQL L++++ ++P+VSIEDPFDQDD+++++ TA +G +
Sbjct 257 YDLDFKSPT-DPSRYITGDQLGALYQDFVRDYPVVSIEDPFDQDDWAAWSKFTANVG--I 313
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+VGDDL VTNP RI +A++ KACN LLLKVNQIGS+TEAI+ACKLAQ +GWGVMVSHRS
Sbjct 314 QIVGDDLTVTNPKRIERAVEEKACNCLLLKVNQIGSVTEAIQACKLAQENGWGVMVSHRS 373
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNPS 186
GETED+FIADLVVGL TGQIKTGAPCRSERL KYNQL+RIEE+L +AG NFRNPS
Sbjct 374 GETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLMRIEEELGDEARFAGHNFRNPS 432
> Hs4503571
Length=434
Score = 264 bits (674), Expect = 8e-71, Method: Compositional matrix adjust.
Identities = 125/178 (70%), Positives = 148/178 (83%), Gaps = 3/178 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFKSP D R ++ DQL DL+K + +++P+VSIEDPFDQDD+ ++ TA G +
Sbjct 257 YDLDFKSPD-DPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDDWGAWQKFTASAG--I 313
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
QVVGDDL VTNP RI KA+ K+CN LLLKVNQIGS+TE+++ACKLAQA+GWGVMVSHRS
Sbjct 314 QVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLAQANGWGVMVSHRS 373
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNP 185
GETED+FIADLVVGL TGQIKTGAPCRSERL KYNQLLRIEE+L + +AG NFRNP
Sbjct 374 GETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSKAKFAGRNFRNP 431
> Hs22064391
Length=377
Score = 264 bits (674), Expect = 8e-71, Method: Compositional matrix adjust.
Identities = 125/178 (70%), Positives = 148/178 (83%), Gaps = 3/178 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFKSP D R ++ DQL DL+K + +++P+VSIEDPFDQDD+ ++ TA G +
Sbjct 200 YDLDFKSPD-DPSRYISPDQLADLYKSFIKDYPVVSIEDPFDQDDWGAWQKFTASAG--I 256
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
QVVGDDL VTNP RI KA+ K+CN LLLKVNQIGS+TE+++ACKLAQA+GWGVMVSHRS
Sbjct 257 QVVGDDLTVTNPKRIAKAVNEKSCNCLLLKVNQIGSVTESLQACKLAQANGWGVMVSHRS 316
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNP 185
GETED+FIADLVVGL TGQIKTGAPCRSERL KYNQLLRIEE+L + +AG NFRNP
Sbjct 317 GETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLLRIEEELGSKAKFAGRNFRNP 374
> CE03684
Length=434
Score = 257 bits (656), Expect = 1e-68, Method: Compositional matrix adjust.
Identities = 120/185 (64%), Positives = 151/185 (81%), Gaps = 3/185 (1%)
Query 1 YSKEAKSYDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALT 60
+ K+ K YDLDFK+PA+D+ + L+G+QL +L++ + +E+P+VSIED FDQDD+ ++
Sbjct 251 FFKDGK-YDLDFKNPASDSSKWLSGEQLTELYQSFIKEYPVVSIEDAFDQDDWDNWGKFH 309
Query 61 AEIGSKVQVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWG 120
+ +Q+VGDDL VTNP RI+ A+ K+CN LLLKVNQIGS+TE+IEA KL++A+GWG
Sbjct 310 G--ATSIQLVGDDLTVTNPKRIQTAIDKKSCNCLLLKVNQIGSVTESIEAAKLSRANGWG 367
Query 121 VMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGE 180
VMVSHRSGETED+FIADLVVGL TGQIKTGAPCRSERL KYNQLLRIEE+L YAG
Sbjct 368 VMVSHRSGETEDTFIADLVVGLATGQIKTGAPCRSERLAKYNQLLRIEEELGADAVYAGH 427
Query 181 NFRNP 185
NFRNP
Sbjct 428 NFRNP 432
> Hs4503573
Length=434
Score = 254 bits (650), Expect = 5e-68, Method: Compositional matrix adjust.
Identities = 119/178 (66%), Positives = 148/178 (83%), Gaps = 3/178 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFKSP D R +TG++L +L+K + + +P+VSIEDPFDQDD++++ + + G +
Sbjct 257 YDLDFKSPD-DPARHITGEKLGELYKSFIKNYPVVSIEDPFDQDDWATWTSFLS--GVNI 313
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+VGDDL VTNP RI +A++ KACN LLLKVNQIGS+TE+I+ACKLAQ++GWGVMVSHRS
Sbjct 314 QIVGDDLTVTNPKRIAQAVEKKACNCLLLKVNQIGSVTESIQACKLAQSNGWGVMVSHRS 373
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNP 185
GETED+FIADLVVGL TGQIKTGAPCRSERL KYNQL+RIEE L + +AG FRNP
Sbjct 374 GETEDTFIADLVVGLCTGQIKTGAPCRSERLAKYNQLMRIEEALGDKAIFAGRKFRNP 431
> At1g74030
Length=477
Score = 243 bits (621), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 116/178 (65%), Positives = 148/178 (83%), Gaps = 3/178 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDL+FK D +L+ + L DL++E+ ++FPIVSIEDPFDQDD+SS+A+L + + +
Sbjct 303 YDLNFKKQPNDGAHVLSAESLADLYREFIKDFPIVSIEDPFDQDDWSSWASLQSSV--DI 360
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+VGDDLLVTNP RI +A++ ++CNALLLKVNQIG++TE+I+A ++A+GWGVMVSHRS
Sbjct 361 QLVGDDLLVTNPKRIAEAIKKQSCNALLLKVNQIGTVTESIQAALDSKAAGWGVMVSHRS 420
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNP 185
GETED+FIADL VGL +GQIKTGAPCRSERL KYNQLLRIEE+L G YAGE FR+P
Sbjct 421 GETEDNFIADLSVGLASGQIKTGAPCRSERLSKYNQLLRIEEEL-GNVRYAGEAFRSP 477
> 7296048
Length=433
Score = 236 bits (603), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 111/178 (62%), Positives = 141/178 (79%), Gaps = 2/178 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFK+ +D + L D+L +L++E+ ++FPIVSIEDPFDQD + +++ LT + +
Sbjct 257 YDLDFKNEKSDKSQWLPADKLANLYQEFIKDFPIVSIEDPFDQDHWEAWSNLTG--CTDI 314
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+VGDDL VTNP RI A++ KACN LLLKVNQIG++TE+I A LA+ +GWG MVSHRS
Sbjct 315 QIVGDDLTVTNPKRIATAVEKKACNCLLLKVNQIGTVTESIAAHLLAKKNGWGTMVSHRS 374
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRNP 185
GETEDSFI DLVVGL TGQIKTGAPCRSERL KYNQ+LRIEE++ +AG++FR P
Sbjct 375 GETEDSFIGDLVVGLSTGQIKTGAPCRSERLAKYNQILRIEEEIGAGVKFAGKSFRKP 432
> YGR254w
Length=437
Score = 228 bits (582), Expect = 4e-60, Method: Compositional matrix adjust.
Identities = 109/177 (61%), Positives = 137/177 (77%), Gaps = 2/177 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFK+P +D + LTG QL DL+ + +PIVSIEDPF +DD+ +++ G +
Sbjct 259 YDLDFKNPNSDKSKWLTGPQLADLYHSLMKRYPIVSIEDPFAEDDWEAWSHFFKTAG--I 316
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+V DDL VTNP RI A++ KA +ALLLKVNQIG+++E+I+A + + A+GWGVMVSHRS
Sbjct 317 QIVADDLTVTNPKRIATAIEKKAADALLLKVNQIGTLSESIKAAQDSFAAGWGVMVSHRS 376
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRN 184
GETED+FIADLVVGLRTGQIKTGAP RSERL K NQLLRIEE+L +AGENF +
Sbjct 377 GETEDTFIADLVVGLRTGQIKTGAPARSERLAKLNQLLRIEEELGDNAVFAGENFHH 433
> YHR174w
Length=437
Score = 228 bits (581), Expect = 5e-60, Method: Compositional matrix adjust.
Identities = 108/177 (61%), Positives = 138/177 (77%), Gaps = 2/177 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLDFK+P +D + LTG +L D++ + +PIVSIEDPF +DD+ +++ G +
Sbjct 259 YDLDFKNPESDKSKWLTGVELADMYHSLMKRYPIVSIEDPFAEDDWEAWSHFFKTAG--I 316
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+V DDL VTNPARI A++ KA +ALLLKVNQIG+++E+I+A + + A+ WGVMVSHRS
Sbjct 317 QIVADDLTVTNPARIATAIEKKAADALLLKVNQIGTLSESIKAAQDSFAANWGVMVSHRS 376
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFRN 184
GETED+FIADLVVGLRTGQIKTGAP RSERL K NQLLRIEE+L + YAGENF +
Sbjct 377 GETEDTFIADLVVGLRTGQIKTGAPARSERLAKLNQLLRIEEELGDKAVYAGENFHH 433
> SPBC1815.01
Length=439
Score = 211 bits (537), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 101/172 (58%), Positives = 128/172 (74%), Gaps = 2/172 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLD K+ LT QL DL+ E S+++PIVSIEDPFDQDD+S++ + AE +
Sbjct 258 YDLDIKAAKPKPENKLTYQQLTDLYVELSKKYPIVSIEDPFDQDDWSAWTHMKAE--TDF 315
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
Q+VGDDL VTN R+R A+ K NALLLKVNQIGS+TE++ A +++ +GWGVMVSHRS
Sbjct 316 QIVGDDLTVTNVKRLRTAIDKKCANALLLKVNQIGSVTESLNAVRMSYEAGWGVMVSHRS 375
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAG 179
GET D+FI+ L VG+ GQ+K+GAPCRSERL KYN+LLRIEE+L YAG
Sbjct 376 GETADTFISHLTVGIGAGQLKSGAPCRSERLAKYNELLRIEEELGSEGVYAG 427
> At2g29560
Length=475
Score = 206 bits (523), Expect = 3e-53, Method: Compositional matrix adjust.
Identities = 98/176 (55%), Positives = 129/176 (73%), Gaps = 2/176 (1%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALTAEIGSKV 67
YDLD KSP + + + D++KE ++PIVSIEDPFD++D+ + + +G
Sbjct 299 YDLDIKSPNKSGQNFKSAEDMIDMYKEICNDYPIVSIEDPFDKEDWE-HTKYFSSLGI-C 356
Query 68 QVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRS 127
QVVGDDLL++N R+ +A+Q +CNALLLKVNQIG++TEAIE K+A+ + WGV+ SHR
Sbjct 357 QVVGDDLLMSNSKRVERAIQESSCNALLLKVNQIGTVTEAIEVVKMARDAQWGVVTSHRC 416
Query 128 GETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGENFR 183
GETEDSFI+DL VGL TG IK GAPCR ER KYNQLLRIEE+L + YAGE+++
Sbjct 417 GETEDSFISDLSVGLATGVIKAGAPCRGERTMKYNQLLRIEEELGDQAVYAGEDWK 472
> Hs7657062
Length=458
Score = 205 bits (522), Expect = 4e-53, Method: Compositional matrix adjust.
Identities = 119/191 (62%), Positives = 142/191 (74%), Gaps = 14/191 (7%)
Query 8 YDLDFKSPAADAHRLLTGDQLKDLFKEWS-----EEFPI-VSIEDP-FDQDDFSSYAALT 60
YDLDF SP D R ++ DQL DL+K + + +P+ VSIEDP FDQDD+ ++ L
Sbjct 266 YDLDFNSPD-DPSRYISPDQLADLYKGFVLGHAVKNYPVGVSIEDPPFDQDDWGAWKKLF 324
Query 61 AEIGSKVQVVGDDLLVTNP-ARIRKALQH-KACNALLL-KVNQIGSITEAIEACKLAQAS 117
+QVVGDDL VT P ARI KA++ KACN LLL KVNQIGS+TE+++ACKLAQ++
Sbjct 325 TGSLVGIQVVGDDLTVTKPEARIAKAVEEVKACNCLLLLKVNQIGSVTESLQACKLAQSN 384
Query 118 GWGVM-VSHR-SGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQG-R 174
GWGVM VSHR SGETED+F+ADLVVGL TGQIKTG CRSERL KYNQLLRIEE G +
Sbjct 385 GWGVMPVSHRLSGETEDTFMADLVVGLCTGQIKTGPTCRSERLAKYNQLLRIEEAEAGSK 444
Query 175 CTYAGENFRNP 185
+AG NFRNP
Sbjct 445 ARFAGRNFRNP 455
> YMR323w
Length=437
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 99/184 (53%), Positives = 132/184 (71%), Gaps = 3/184 (1%)
Query 1 YSKEAKSYDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALT 60
+ K+ K YDL+FK P +D L+ QL + + +++PI+S+EDP+ +DD+SS++A
Sbjct 253 FYKDGK-YDLNFKEPNSDPSHWLSPAQLAEYYHSLLKKYPIISLEDPYAEDDWSSWSAFL 311
Query 61 AEIGSKVQVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWG 120
+ VQ++ DDL TN RI +A++ K N LLLK+NQIG++TE+IEA A +GWG
Sbjct 312 KTV--NVQIIADDLTCTNKTRIARAIEEKCANTLLLKLNQIGTLTESIEAANQAFDAGWG 369
Query 121 VMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGE 180
VM+SHRSGETED FIADLVVGLR GQIK+GA RSERL KYN+LLRIEE+L C YAG
Sbjct 370 VMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGDDCIYAGH 429
Query 181 NFRN 184
F +
Sbjct 430 RFHD 433
> YPL281c
Length=437
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 99/184 (53%), Positives = 132/184 (71%), Gaps = 3/184 (1%)
Query 1 YSKEAKSYDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALT 60
+ K+ K YDL+FK P +D L+ QL + + +++PI+S+EDP+ +DD+SS++A
Sbjct 253 FYKDGK-YDLNFKEPNSDPSHWLSPAQLAEYYHSLLKKYPIISLEDPYAEDDWSSWSAFL 311
Query 61 AEIGSKVQVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWG 120
+ VQ++ DDL TN RI +A++ K N LLLK+NQIG++TE+IEA A +GWG
Sbjct 312 KTV--NVQIIADDLTCTNKTRIARAIEEKCANTLLLKLNQIGTLTESIEAANQAFDAGWG 369
Query 121 VMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGE 180
VM+SHRSGETED FIADLVVGLR GQIK+GA RSERL KYN+LLRIEE+L C YAG
Sbjct 370 VMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGDDCIYAGH 429
Query 181 NFRN 184
F +
Sbjct 430 RFHD 433
> YOR393w
Length=437
Score = 203 bits (516), Expect = 2e-52, Method: Compositional matrix adjust.
Identities = 99/184 (53%), Positives = 132/184 (71%), Gaps = 3/184 (1%)
Query 1 YSKEAKSYDLDFKSPAADAHRLLTGDQLKDLFKEWSEEFPIVSIEDPFDQDDFSSYAALT 60
+ K+ K YDL+FK P +D L+ QL + + +++PI+S+EDP+ +DD+SS++A
Sbjct 253 FYKDGK-YDLNFKEPNSDPSHWLSPAQLAEYYHSLLKKYPIISLEDPYAEDDWSSWSAFL 311
Query 61 AEIGSKVQVVGDDLLVTNPARIRKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWG 120
+ VQ++ DDL TN RI +A++ K N LLLK+NQIG++TE+IEA A +GWG
Sbjct 312 KTV--NVQIIADDLTCTNKTRIARAIEEKCANTLLLKLNQIGTLTESIEAANQAFDAGWG 369
Query 121 VMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQLLRIEEQLQGRCTYAGE 180
VM+SHRSGETED FIADLVVGLR GQIK+GA RSERL KYN+LLRIEE+L C YAG
Sbjct 370 VMISHRSGETEDPFIADLVVGLRCGQIKSGALSRSERLAKYNELLRIEEELGDDCIYAGH 429
Query 181 NFRN 184
F +
Sbjct 430 RFHD 433
> ECU10g1690
Length=412
Score = 155 bits (392), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 72/147 (48%), Positives = 103/147 (70%), Gaps = 1/147 (0%)
Query 24 TGDQLKDLFKEWSEEFP-IVSIEDPFDQDDFSSYAALTAEIGSKVQVVGDDLLVTNPARI 82
T L + + E +E+P + S+EDPF + D+ + L AE+G K+ +VGDDL VT+P +
Sbjct 265 TTKSLGERYIEILKEYPQVYSLEDPFSERDYDGWIWLNAEVGKKINIVGDDLTVTDPQLV 324
Query 83 RKALQHKACNALLLKVNQIGSITEAIEACKLAQASGWGVMVSHRSGETEDSFIADLVVGL 142
R A + CN LL+K NQ+G+++E +EA ++A+ G +MVSHRSGET+D FI+DL VG+
Sbjct 325 RDAGARRMCNTLLVKPNQVGTVSETVEAIRIARKCGMKIMVSHRSGETDDHFISDLSVGV 384
Query 143 RTGQIKTGAPCRSERLCKYNQLLRIEE 169
IK+GAPCR ER+ KYNQLLR+ E
Sbjct 385 GAEYIKSGAPCRGERVSKYNQLLRLYE 411
> Hs18575411
Length=98
Score = 46.6 bits (109), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 27/75 (36%), Positives = 38/75 (50%), Gaps = 0/75 (0%)
Query 104 ITEAIEACKLAQASGWGVMVSHRSGETEDSFIADLVVGLRTGQIKTGAPCRSERLCKYNQ 163
+++ +E L + + GE+ D + DL VGL IK G R ER+ KYN+
Sbjct 1 MSDLVEITNLIDSKKHITVFGSTEGESSDDSLVDLAVGLGVRFIKLGGLSRGERVTKYNR 60
Query 164 LLRIEEQLQGRCTYA 178
LL IEE+L T A
Sbjct 61 LLTIEEELVQNGTLA 75
Lambda K H
0.316 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3058524910
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40