bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4093_orf1
Length=184
Score E
Sequences producing significant alignments: (Bits) Value
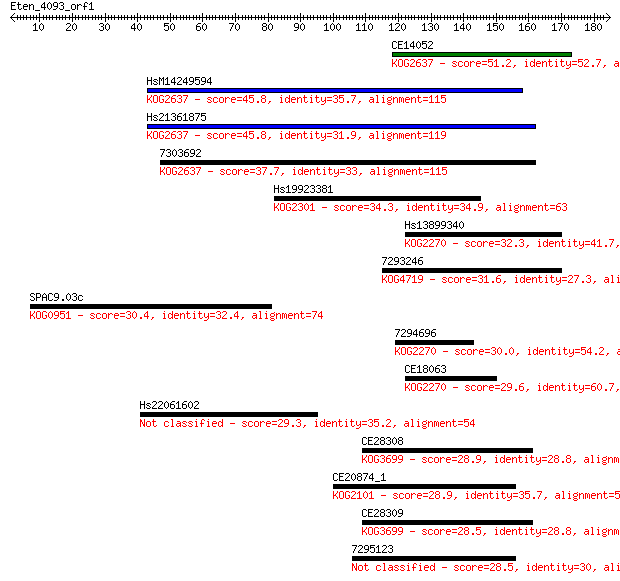
CE14052 51.2 1e-06
HsM14249594 45.8 5e-05
Hs21361875 45.8 5e-05
7303692 37.7 0.011
Hs19923381 34.3 0.14
Hs13899340 32.3 0.61
7293246 31.6 1.1
SPAC9.03c 30.4 2.3
7294696 30.0 2.8
CE18063 29.6 3.3
Hs22061602 29.3 4.9
CE28308 28.9 6.5
CE20874_1 28.9 6.8
CE28309 28.5 6.9
7295123 28.5 8.6
> CE14052
Length=410
Score = 51.2 bits (121), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 29/56 (51%), Positives = 38/56 (67%), Gaps = 1/56 (1%)
Query 118 VSTYRPRGETTEERKARKTAVKEAQRLLRANKKANKQLLKEETKN-AQARQARISA 172
VST+RP+ ET E+R RK AVKEA++L R KKANK + EE + A+ R +I A
Sbjct 352 VSTFRPKNETPEQRSLRKKAVKEARKLRRVEKKANKTMFAEEKRKLAKGRIGQIKA 407
> HsM14249594
Length=299
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 41/124 (33%), Positives = 58/124 (46%), Gaps = 23/124 (18%)
Query 43 ARPSWDGETIISARSGVSVQPRSI--------IVGSSRTTKLSLTPYLALGGGPLGVKGP 94
A+ WD E+I S S + P+ I I SS+T G PL V
Sbjct 168 AKEKWDCESICSTYSNLYNHPQLIKYQPKPKQIRISSKT------------GIPLNVLPK 215
Query 95 QGPRRAQLPPTLEETGTAVELPEVSTY-RPRGETTEERKARKTAVKEAQRLLRANKKANK 153
+G Q G+ +LP+VST R + E+ E+++ARK A+KE ++ R KKANK
Sbjct 216 KGLTAKQTERIQMINGS--DLPKVSTQPRSKNESKEDKRARKQAIKEERKERRVEKKANK 273
Query 154 QLLK 157
K
Sbjct 274 LAFK 277
> Hs21361875
Length=475
Score = 45.8 bits (107), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 38/120 (31%), Positives = 62/120 (51%), Gaps = 7/120 (5%)
Query 43 ARPSWDGETIISARSGVSVQPRSIIVGSSRTTKLSLTPYLALGGGPLGVKGPQGPRRAQL 102
A+ WD E+I S S + P+ +I + ++ ++ + G PL V +G Q
Sbjct 344 AKEKWDCESICSTYSNLYNHPQ-LIKYQPKPKQIRIS---SKTGIPLNVLPKKGLTAKQT 399
Query 103 PPTLEETGTAVELPEVSTY-RPRGETTEERKARKTAVKEAQRLLRANKKANKQLLKEETK 161
G+ +LP+VST R + E+ E+++ARK A+KE ++ R KKANK K E +
Sbjct 400 ERIQMINGS--DLPKVSTQPRSKNESKEDKRARKQAIKEERKERRVEKKANKLAFKLEKR 457
> 7303692
Length=493
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 55/135 (40%), Gaps = 21/135 (15%)
Query 47 WDGETIISARSGVSVQPRSI--------IVGSSRTTKLSLTPYLALGGGPLGVKGPQGPR 98
WD E+I+S S + P+ I S+ + + P L L G G
Sbjct 341 WDCESILSTYSNIYNHPKVIDEPRRSRRSSASTNPAPIQIDPKTGLPTNVLR-GGVDGQL 399
Query 99 RAQLPPTLEETGTAVELPE------------VSTYRPRGETTEERKARKTAVKEAQRLLR 146
A+ L + A P+ V + RP+ ET EE+K RK +K+ + R
Sbjct 400 TAKALANLADESPAATGPKSLCAKSVLSTLSVLSIRPKDETHEEKKERKRLLKDYRNERR 459
Query 147 ANKKANKQLLKEETK 161
KKAN + KEE K
Sbjct 460 IEKKANTEAFKEEKK 474
> Hs19923381
Length=2000
Score = 34.3 bits (77), Expect = 0.14, Method: Composition-based stats.
Identities = 22/63 (34%), Positives = 30/63 (47%), Gaps = 4/63 (6%)
Query 82 LALGGGPLGVKGPQGPRRAQLPPTLEETGTAVELPEVSTYRPRGETTEERKARKTAVKEA 141
++L GGP + P G QLPP T T V +S+Y+ E E+ R+ AV A
Sbjct 654 VSLVGGPSALTSPTG----QLPPEGTTTETEVRKRRLSSYQISMEMLEDSSGRQRAVSIA 709
Query 142 QRL 144
L
Sbjct 710 SIL 712
> Hs13899340
Length=561
Score = 32.3 bits (72), Expect = 0.61, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 30/51 (58%), Gaps = 4/51 (7%)
Query 122 RPRGETTE---ERKARKTAVKEAQRLLRANKKANKQLLKEETKNAQARQAR 169
RP+ TT+ ++K RK VKEAQR R N K K + K + K A+ ++ +
Sbjct 512 RPKKHTTDPDIDKKERKKMVKEAQREKRKN-KIPKHVKKRKEKTAKTKKGK 561
> 7293246
Length=1836
Score = 31.6 bits (70), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 15/55 (27%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 115 LPEVSTYRPRGETTEERKARKTAVKEAQRLLRANKKANKQLLKEETKNAQARQAR 169
P ++ P G + RK + E Q L NK ++++ + +T N+QAR +
Sbjct 1106 YPSINNNLPAGSGFDISFTRKANMWECQTCLVMNKSSDEECIACQTPNSQARNSN 1160
> SPAC9.03c
Length=2176
Score = 30.4 bits (67), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 15/85 (17%)
Query 7 DIDDELRDK-----------TLALARLQEEEEERAPTVHVEPLLPARARPSWDGETIISA 55
DID E+ D + L R EE+EE TV + P PA+ W ++ +
Sbjct 2054 DIDFEIEDSEDVHANSPSVLIVQLTRELEEDEEVDTTV-IAPYFPAQKTEHW---WLVIS 2109
Query 56 RSGVSVQPRSIIVGSSRTTKLSLTP 80
+ + I +G S TTK+ P
Sbjct 2110 DDKTLLAIKKITLGRSLTTKMEFVP 2134
> 7294696
Length=585
Score = 30.0 bits (66), Expect = 2.8, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 119 STYRPRGETTEERKARKTAVKEAQ 142
++ RPR E+ E +KARK AVK+A+
Sbjct 538 NSARPREESPESKKARKKAVKDAK 561
> CE18063
Length=548
Score = 29.6 bits (65), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 17/28 (60%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 122 RPRGETTEERKARKTAVKEAQRLLRANK 149
R R ET EERK RK AVKE +R R K
Sbjct 506 RNREETAEERKERKAAVKEEKREQRKEK 533
> Hs22061602
Length=1246
Score = 29.3 bits (64), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 28/64 (43%), Gaps = 12/64 (18%)
Query 41 ARARPSWDGETIISARSGVSVQPRSIIVGSSRTTKLSLTPYLALGG----------GPLG 90
R P W G + G + RS+ VG +RT+ + +L++G G LG
Sbjct 438 GRCYPKWVGNCKVDG--GAGAEGRSLAVGCARTSWMPCGYFLSVGIWAQVYRWLLLGTLG 495
Query 91 VKGP 94
KGP
Sbjct 496 AKGP 499
> CE28308
Length=679
Score = 28.9 bits (63), Expect = 6.5, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 109 TGTAVELPEVSTYRPRGETTEERKARKTAVKEAQRLLRANKKANKQLLKEET 160
+GT V++ V + P +E K ++ A+KE RL + Q+L T
Sbjct 464 SGTPVKISSVHQFSPASSNPKEFKEKQKAIKENNRLGTLSAGPQSQILDSVT 515
> CE20874_1
Length=168
Score = 28.9 bits (63), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 32/58 (55%), Gaps = 5/58 (8%)
Query 100 AQLPPTLEETGTAVELPEVSTYRPRGETT--EERKARKTAVKEAQRLLRANKKANKQL 155
A L PT E+TG + P+V+ ET+ E+R ARK+++K LR + N ++
Sbjct 112 ALLEPTHEQTGDDFQFPDVAI---ASETSNEEQRTARKSSMKRLFPRLRGSTHHNIEI 166
> CE28309
Length=732
Score = 28.5 bits (62), Expect = 6.9, Method: Composition-based stats.
Identities = 15/52 (28%), Positives = 25/52 (48%), Gaps = 0/52 (0%)
Query 109 TGTAVELPEVSTYRPRGETTEERKARKTAVKEAQRLLRANKKANKQLLKEET 160
+GT V++ V + P +E K ++ A+KE RL + Q+L T
Sbjct 464 SGTPVKISSVHQFSPASSNPKEFKEKQKAIKENNRLGTLSAGPQSQILDSVT 515
> 7295123
Length=240
Score = 28.5 bits (62), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 31/50 (62%), Gaps = 0/50 (0%)
Query 106 LEETGTAVELPEVSTYRPRGETTEERKARKTAVKEAQRLLRANKKANKQL 155
L++T A+ Y+P+G+T + K + T +K++ R+ R +KKA +++
Sbjct 60 LQKTHLAITRGSYIVYKPKGKTRLDPKKKITRLKKSVRVYRTSKKAEREV 109
Lambda K H
0.314 0.132 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2950576972
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40