bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4121_orf1
Length=111
Score E
Sequences producing significant alignments: (Bits) Value
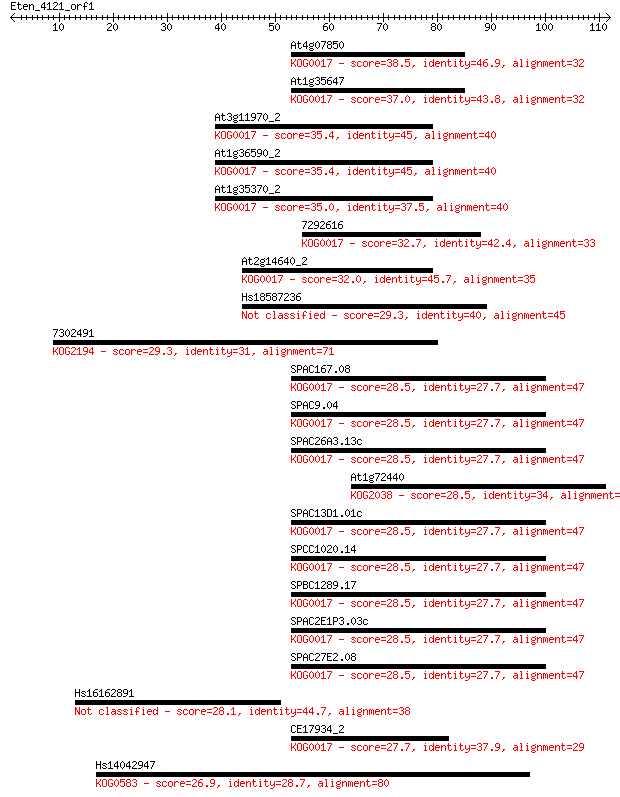
At4g07850 38.5 0.003
At1g35647 37.0 0.010
At3g11970_2 35.4 0.029
At1g36590_2 35.4 0.030
At1g35370_2 35.0 0.033
7292616 32.7 0.18
At2g14640_2 32.0 0.32
Hs18587236 29.3 1.7
7302491 29.3 1.7
SPAC167.08 28.5 3.4
SPAC9.04 28.5 3.4
SPAC26A3.13c 28.5 3.4
At1g72440 28.5 3.4
SPAC13D1.01c 28.5 3.5
SPCC1020.14 28.5 3.6
SPBC1289.17 28.5 3.6
SPAC2E1P3.03c 28.5 3.6
SPAC27E2.08 28.5 3.6
Hs16162891 28.1 4.4
CE17934_2 27.7 6.2
Hs14042947 26.9 10.0
> At4g07850
Length=1138
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 24/32 (75%), Gaps = 0/32 (0%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANL 84
+E AYN + HS+T+ SPF+++ G NP+T +L
Sbjct 961 VEFAYNHSVHSATKFSPFQIVYGFNPITPLDL 992
> At1g35647
Length=1495
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 14/32 (43%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANL 84
+E AYN + HS+++ SPF+++ G NP T +L
Sbjct 1194 VEFAYNHSMHSASKFSPFQIVYGFNPTTPLDL 1225
> At3g11970_2
Length=958
Score = 35.4 bits (80), Expect = 0.029, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Query 39 CHGRLPQQDGSFRSL-ELAYNTTSHSSTELSPFEVMTGENP 78
CH R PQ + +L E YNT HSS+ ++PFE++ G+ P
Sbjct 728 CHDR-PQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVP 767
> At1g36590_2
Length=958
Score = 35.4 bits (80), Expect = 0.030, Method: Composition-based stats.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 2/41 (4%)
Query 39 CHGRLPQQDGSFRSL-ELAYNTTSHSSTELSPFEVMTGENP 78
CH R PQ + +L E YNT HSS+ ++PFE++ G+ P
Sbjct 728 CHDR-PQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVP 767
> At1g35370_2
Length=923
Score = 35.0 bits (79), Expect = 0.033, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 39 CHGRLPQQDGSFRSLELAYNTTSHSSTELSPFEVMTGENP 78
CH R + E YNT HSS++++PFE++ G+ P
Sbjct 716 CHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAP 755
> 7292616
Length=1062
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 55 LAYNTTSHSSTELSPFEVMTGENPLTAANLDIV 87
+ YN HSST+L+P+EV TG + N V
Sbjct 930 ITYNNAIHSSTKLTPYEVFTGRTHIFEQNYKAV 962
> At2g14640_2
Length=492
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 16/36 (44%), Positives = 22/36 (61%), Gaps = 1/36 (2%)
Query 44 PQQDGSFRSL-ELAYNTTSHSSTELSPFEVMTGENP 78
P+Q SF E YNTT H+ST ++PF+ + G P
Sbjct 256 PKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRPP 291
> Hs18587236
Length=466
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Query 44 PQQDGSFRSLELAYNTTSHSSTELS-----PFEVMTGENPLTAANLDIVG 88
PQ D S + ELA + HS E+S F + G+ PLT + D+ G
Sbjct 269 PQLDDSTVARELAITDSEHSDAEVSCTDNGTFNLSRGQTPLTEGSEDLDG 318
> 7302491
Length=891
Score = 29.3 bits (64), Expect = 1.7, Method: Composition-based stats.
Identities = 22/74 (29%), Positives = 35/74 (47%), Gaps = 6/74 (8%)
Query 9 VAHPFSSVGACESLIYHRYSPYNDRARLDSCHGRLPQQDGSFRSLELAYNTTSHSSTELS 68
+ HP++S E L H + P + R+ HG +P D ++ Y+ T H E+
Sbjct 289 IKHPYASTMG-EELFQHNFIPSDTDFRIFRDHGSVPGLDMAYTYNGFVYH-TRHDKAEIF 346
Query 69 P---FEVMTGENPL 79
P F+ TG+N L
Sbjct 347 PRGSFQ-HTGDNLL 359
> SPAC167.08
Length=1214
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 995 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1031
> SPAC9.04
Length=1333
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> SPAC26A3.13c
Length=1333
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> At1g72440
Length=1056
Score = 28.5 bits (62), Expect = 3.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 30/47 (63%), Gaps = 5/47 (10%)
Query 64 STELSPFEVMTGENPLTAANLDIVGALAPTLTPAMTKLFRQLAFQQL 110
+ +++ FE+M GENP+ AN+ + AL +T +K+ ++ AF+ L
Sbjct 214 ADKITAFEIMVGENPI--ANMRSLDALLGMVT---SKVGKRFAFKGL 255
> SPAC13D1.01c
Length=1333
Score = 28.5 bits (62), Expect = 3.5, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> SPCC1020.14
Length=1333
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> SPBC1289.17
Length=1333
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> SPAC2E1P3.03c
Length=1333
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> SPAC27E2.08
Length=1333
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 13/47 (27%), Positives = 26/47 (55%), Gaps = 10/47 (21%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTAANLDIVGALAPTLTPAMT 99
++ +YN HS+T+++PFE++ +P AL+P P+ +
Sbjct 1114 VQQSYNNAIHSATQMTPFEIVHRYSP----------ALSPLELPSFS 1150
> Hs16162891
Length=138
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Query 13 FSSVGACESL-IYHRYSPYNDRARLDSCHGRLPQQDGSF 50
SSV E L + R SP+NDRA+L S + Q G F
Sbjct 87 LSSVLPPEGLSVLGRKSPFNDRAQLRSVREQRKQASGRF 125
> CE17934_2
Length=696
Score = 27.7 bits (60), Expect = 6.2, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 18/29 (62%), Gaps = 0/29 (0%)
Query 53 LELAYNTTSHSSTELSPFEVMTGENPLTA 81
+ YNTT H +T +PF ++ G +P+ A
Sbjct 470 MTFVYNTTVHDTTGETPFFLIFGRDPVFA 498
> Hs14042947
Length=367
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 31/82 (37%), Gaps = 2/82 (2%)
Query 17 GACESLIYHRYSP-YNDRARLDSCHGRLPQQDGSFRSLELAYNTTSHSSTELSP-FEVMT 74
G C+ LIYH P N R +D Q + S +A N SS P +
Sbjct 243 GECKDLIYHMLQPDVNRRLHIDEILSHCWMQPKARGSPSVAINKEGESSRGTEPLWTPEP 302
Query 75 GENPLTAANLDIVGALAPTLTP 96
G + +A L+ G P P
Sbjct 303 GSDKKSATKLEPEGEAQPQAQP 324
Lambda K H
0.318 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40