bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4192_orf1
Length=267
Score E
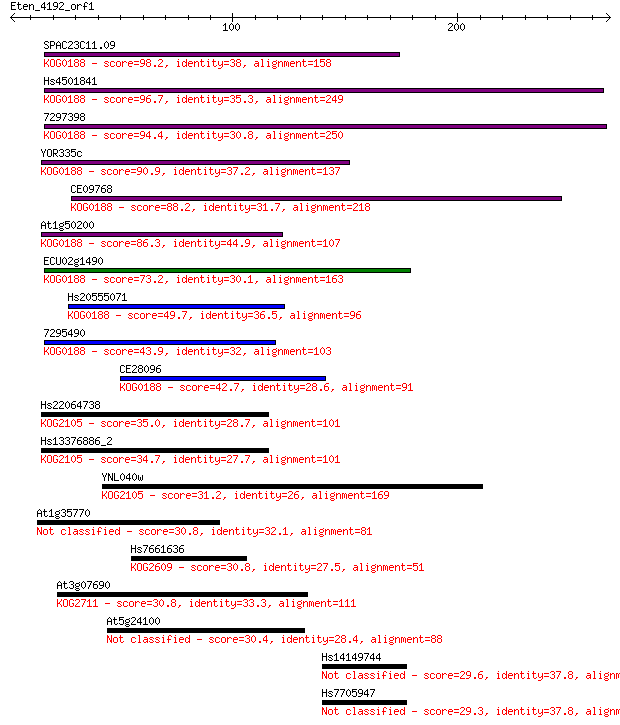
Sequences producing significant alignments: (Bits) Value
SPAC23C11.09 98.2 2e-20
Hs4501841 96.7 4e-20
7297398 94.4 3e-19
YOR335c 90.9 3e-18
CE09768 88.2 2e-17
At1g50200 86.3 6e-17
ECU02g1490 73.2 6e-13
Hs20555071 49.7 7e-06
7295490 43.9 3e-04
CE28096 42.7 7e-04
Hs22064738 35.0 0.18
Hs13376886_2 34.7 0.21
YNL040w 31.2 2.1
At1g35770 30.8 2.7
Hs7661636 30.8 3.2
At3g07690 30.8 3.5
At5g24100 30.4 3.6
Hs14149744 29.6 6.9
Hs7705947 29.3 8.4
> SPAC23C11.09
Length=959
Score = 98.2 bits (243), Expect = 2e-20, Method: Composition-based stats.
Identities = 60/160 (37%), Positives = 85/160 (53%), Gaps = 9/160 (5%)
Query 16 IASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVS 75
I +L V + + L A E+ L AVFGE YPDPVRVV IG D L +QE + D
Sbjct 662 IQDNLSVYSKEVALSKAKEINGLRAVFGEVYPDPVRVVCIGVDIDTL--LQEPKKPDWTK 719
Query 76 SSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISARSELRDVCLSLSNL 135
S+E CGGTH + +++DFV++ E IAKG+RRIVAV A + ++ L
Sbjct 720 YSIEFCGGTHCDKSGEIKDFVILEENGIAKGIRRIVAVTSTEANRVSRLANEFDARIAKL 779
Query 136 ESA--SPAFPFFEKEVQQAKAFVESNKMIPLLSRRRLLAD 173
E SPA KE + K V+ +K++ R+ + +
Sbjct 780 EKMPFSPA-----KEAELKKISVDLSKLVVAAVRKHAMKE 814
> Hs4501841
Length=968
Score = 96.7 bits (239), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 88/263 (33%), Positives = 127/263 (48%), Gaps = 29/263 (11%)
Query 16 IASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVS 75
I ++ V T PL A + L AVF E YPDPVRVV+IG EL + + G
Sbjct 660 IEAAKAVYTQDCPLAAAKAIQGLRAVFDETYPDPVRVVSIGVPVSEL--LDDPSGPAGSL 717
Query 76 SSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISA--RSELRDVCLSLS 133
+SVE CGGTHL N++ FV+++EE+IAKG+RRIVAV G A A ++E CLS+
Sbjct 718 TSVEFCGGTHLRNSSHAGAFVIVTEEAIAKGIRRIVAVTGAEAQKALRKAESLKKCLSVM 777
Query 134 NLESASPAFPFFEKEVQQAKAFVE---SNKMIPLLSRRRLLADLENKQKLGLELGKFRAK 190
+ + P K+VQ+ A + + +IP + L L++ +K+
Sbjct 778 EAKVKAQTAP--NKDVQREIADLGEALATAVIPQWQKDELRETLKSLKKV--------MD 827
Query 191 ELLRVGKLSGQEAAAAAAAEGKKFIVLN-------LEQLQG-DGKALDEAAKALQSAAPS 242
+L R K Q+ K+FI N LE G KAL+EA K + +P
Sbjct 828 DLDRASKADVQKRVLEKT---KQFIDSNPNQPLVILEMESGASAKALNEALKLFKMHSPQ 884
Query 243 LPFLLLTXLESRGS-SCIXLSPR 264
+L T G +C+ P+
Sbjct 885 TSAMLFTVDNEAGKITCLCQVPQ 907
> 7297398
Length=966
Score = 94.4 bits (233), Expect = 3e-19, Method: Composition-based stats.
Identities = 77/263 (29%), Positives = 131/263 (49%), Gaps = 25/263 (9%)
Query 16 IASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVS 75
+ ++P+ L A ++ L +VF E YPDPVRV++ G EL++ +++ ++
Sbjct 660 VYKNVPIYAKESKLALAKKIRGLRSVFDEVYPDPVRVISFGVPVDELEQNPDSEAGEQ-- 717
Query 76 SSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISA-------RSELRDV 128
+SVE CGGTHL + + DFV+ SEE+IAKG+RRIVA+ G A+ A E+ +
Sbjct 718 TSVEFCGGTHLRRSGHIMDFVISSEEAIAKGIRRIVALTGPEALKALKKSEAFEQEIVRL 777
Query 129 CLSLSNLESASPAFPFFEKEVQQAKAFVESNKMIPLLSR---RRLLADLENKQKLGLELG 185
++ N +S + ++ V+ + S+ IP + + R LL L K+ L +
Sbjct 778 KATIDNDKSGKDSKSHVKEIVELTEQI--SHATIPYVKKDEMRNLLKGL--KKTLDDKER 833
Query 186 KFRAKELLRVGKLSGQEAAAAAAAEGKKFIVLNLEQLQG--DGKALDEAAKALQSAAPSL 243
RA + V + A E + +EQL+ + KALD A K ++S P
Sbjct 834 ALRAAVSVTVVE------RAKTLCEANPNATVLVEQLEAFNNTKALDAALKQVRSQLPDA 887
Query 244 PFLLLTX-LESRGSSCIXLSPRA 265
+ L+ +S+ C+ P++
Sbjct 888 AAMFLSVDADSKKIFCLSSVPKS 910
> YOR335c
Length=958
Score = 90.9 bits (224), Expect = 3e-18, Method: Composition-based stats.
Identities = 51/137 (37%), Positives = 74/137 (54%), Gaps = 3/137 (2%)
Query 15 QIASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKV 74
QI +L V +PL A + + AVFGE YPDPVRVV++G +EL + +
Sbjct 655 QIKENLQVFYKEIPLDLAKSIDGVRAVFGETYPDPVRVVSVGKPIEEL--LANPANEEWT 712
Query 75 SSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISARSELRDVCLSLSN 134
S+E CGGTH++ ++ FV++ E IAKG+RRIVAV G A A+ L
Sbjct 713 KYSIEFCGGTHVNKTGDIKYFVILEESGIAKGIRRIVAVTGTEAFEAQRLAEQFAADLDA 772
Query 135 LESASPAFPFFEKEVQQ 151
+ P P EK++++
Sbjct 773 ADKL-PFSPIKEKKLKE 788
> CE09768
Length=968
Score = 88.2 bits (217), Expect = 2e-17, Method: Composition-based stats.
Identities = 69/229 (30%), Positives = 114/229 (49%), Gaps = 18/229 (7%)
Query 28 PLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVSSSVELCGGTHLS 87
PL A ++ L A+F E YPDPVRVV +G ++L +Q + +++VE CGGTHL
Sbjct 673 PLGEAKKVKGLRAMFDETYPDPVRVVAVGTPVEQL--LQNPDAEEGQNTTVEFCGGTHLQ 730
Query 88 NAAQLEDFVVISEESIAKGVRRIVAVAGEAAISARSELRDVCLSLS----------NLES 137
N + + V+ SEE+IAKG+RRIVA+ G A A + + L L +
Sbjct 731 NVSHIGRIVIASEEAIAKGIRRIVALTGPEAERAIARADRLTARLEEESKHADKKDELLA 790
Query 138 ASPAFPFFEKEVQQAKAFVESN-KMIPLLSRRRLLADLENKQKLGLELGKFRAKELLRVG 196
F +K++Q+ E+N +P + + + QK L+ G +A++
Sbjct 791 NKDKFKALQKKIQE--IVDEANGAQLPYWRKDSIREKAKAIQKT-LD-GYTKAQQAAVAE 846
Query 197 KLSGQEAAAAAAAEGKKFIVLNLEQLQGDGKALDEAAKALQSAAPSLPF 245
K+ G+ AA AE + +++++ + KA+D A K L+ + F
Sbjct 847 KVLGEAKELAAVAE-QPTVLVHVFAANANSKAIDNALKLLKDTKAVMAF 894
> At1g50200
Length=1027
Score = 86.3 bits (212), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 48/107 (44%), Positives = 62/107 (57%), Gaps = 2/107 (1%)
Query 15 QIASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKV 74
QI L V + L A + L AVFGE YPDPVRVV+IG ++L E +
Sbjct 725 QIKDELDVFSKEAVLSEAKRIKGLRAVFGEVYPDPVRVVSIGRKVEDLLADPENNEWSLL 784
Query 75 SSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISA 121
SS E CGGTH++N + + F ++SEE IAKG+RR+ AV E A A
Sbjct 785 SS--EFCGGTHITNTREAKAFALLSEEGIAKGIRRVTAVTTECAFDA 829
> ECU02g1490
Length=864
Score = 73.2 bits (178), Expect = 6e-13, Method: Composition-based stats.
Identities = 49/180 (27%), Positives = 85/180 (47%), Gaps = 33/180 (18%)
Query 16 IASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVS 75
I V +L R AM+ P + + E YP+ VR++T+ D +Q
Sbjct 606 IKKGAEVKAVILDRRQAMDNPNIVRMQNEEYPENVRIITMQSDGLIIQ------------ 653
Query 76 SSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAA--ISARSELRDVCLSLS 133
E+CGGTH++N + + F ++SE ++ RRIV V G+ A + A +EL LS+
Sbjct 654 ---EMCGGTHVTNVSDIGKFRIVSETGVSANTRRIVGVTGKKAHELDAEAELCLSKLSMG 710
Query 134 NLESASPAFPFF---------------EKEVQQAKAFVESNKMIPLLSRRRLLADLENKQ 178
N+ + A P F ++E+ +A+ F + + LL++ + +D N+Q
Sbjct 711 NVVNVDKAIPLFYRQKIEVLNQRNRKKDEELSKAQYFKNKKEFLELLAKAKNASD-SNEQ 769
> Hs20555071
Length=985
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 35/96 (36%), Positives = 53/96 (55%), Gaps = 4/96 (4%)
Query 27 LPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGADKVSSSVELCGGTHL 86
+PL ++P L ++ E YPDPVRVV++ + + + +SVELC GTHL
Sbjct 699 VPLALTAQVPGLRSL-DEVYPDPVRVVSV---GVPVAHALDPASQAALQTSVELCCGTHL 754
Query 87 SNAAQLEDFVVISEESIAKGVRRIVAVAGEAAISAR 122
+ D V+I + ++KG R++AV GE A AR
Sbjct 755 LRTGAVGDLVIIGDRQLSKGTTRLLAVTGEQAQQAR 790
> 7295490
Length=1012
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 52/104 (50%), Gaps = 14/104 (13%)
Query 16 IASSLPVNTAVLPLRTAMELPALCAVFGEAYPDP-VRVVTIGPDTKELQKIQETQGADKV 74
I S+ PV +L +E + V GE YP+ +R+V + ++ ELQ
Sbjct 713 ICSAAPVEVQLLSAAEVLEQNDITMVPGEVYPEQGLRLVNV--ESPELQ----------- 759
Query 75 SSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAGEAA 118
SS ELC GTH +N ++L F +++ + + AVAG+AA
Sbjct 760 LSSKELCCGTHATNTSELSCFCIVNLKQTNRARFAFTAVAGQAA 803
> CE28096
Length=793
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 46/91 (50%), Gaps = 18/91 (19%)
Query 50 VRVVTIGPDTKELQKIQETQGADKVSSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRR 109
VRVV +G GAD VE C GTH+ + ++D ++S++S+ + +RR
Sbjct 690 VRVVALGS------------GAD---VPVECCSGTHIHDVRVIDDVAIMSDKSMGQRLRR 734
Query 110 IVAVAGEAAISARSELRDVCLSLSNLESASP 140
I+ + G+ A + R+ + + +L S P
Sbjct 735 IIVLTGKEAAACRNYTKSI---YEDLRSTDP 762
> Hs22064738
Length=307
Score = 35.0 bits (79), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 26/105 (24%)
Query 15 QIASSLPVNTAVLPLRTAMELPALCAVFGEAYPD----PVRVVTIGPDTKELQKIQETQG 70
+I LPVN L L + P + V G PD P+RVV I +G
Sbjct 56 KIRDRLPVNVRELSL----DDPEVEQVSGRGLPDDHAGPIRVVNI-------------EG 98
Query 71 ADKVSSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAG 115
D +C GTH+SN + L+ ++ E K ++ ++G
Sbjct 99 VDS-----NMCCGTHVSNLSDLQVIKILGTEKGKKNRTNLIFLSG 138
> Hs13376886_2
Length=428
Score = 34.7 bits (78), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 28/105 (26%), Positives = 45/105 (42%), Gaps = 26/105 (24%)
Query 15 QIASSLPVNTAVLPLRTAMELPALCAVFGEAYPD----PVRVVTIGPDTKELQKIQETQG 70
+I LPVN L +++ P + V G PD P+RVV I +G
Sbjct 177 KIRDRLPVNVREL----SLDDPEVEQVSGRGLPDDHAGPIRVVNI-------------EG 219
Query 71 ADKVSSSVELCGGTHLSNAAQLEDFVVISEESIAKGVRRIVAVAG 115
D +C GTH+SN + L+ ++ E K ++ ++G
Sbjct 220 VDS-----NMCCGTHVSNLSDLQVIKILGTEKGKKNRTNLIFLSG 259
> YNL040w
Length=456
Score = 31.2 bits (69), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 44/172 (25%), Positives = 74/172 (43%), Gaps = 23/172 (13%)
Query 42 FGEAYPDPVRVVTIGPDTKELQK-IQETQGADKVSSSVELCGGTHLSNAAQLEDFVVISE 100
GE D V I PD +L K + T + S+ C GTHL +Q+ +++S
Sbjct 203 IGEETVDEVSTSKI-PDDYDLSKGVLRTIHIGDIDSNP--CCGTHLKCTSQIGSILILSN 259
Query 101 ESIAKGVR-RIVAVAGE-AAISARSELRDVCLSLSNLESASPAFPFFEKEVQQAKAFVES 158
+S +G R+ + G+ ++ A+S + + L NL S S E Q ++
Sbjct 260 QSAVRGSNSRLYFMCGKRVSLYAKS-VNKILLDSKNLLSCS--------ETQISEKITRQ 310
Query 159 NKMIPLLSRRRLLADLENKQKLGLELGKFRAKELLRVGKLSGQEAAAAAAAE 210
K I L++R +Q L + ++EL+ K SG++ A E
Sbjct 311 TKQIQQLNKR--------EQYWIKRLARTASEELMNTLKASGKKRAYFMEEE 354
> At1g35770
Length=968
Score = 30.8 bits (68), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 26/81 (32%), Positives = 38/81 (46%), Gaps = 8/81 (9%)
Query 13 LFQIASSLPVNTAVLPLRTAMELPALCAVFGEAYPDPVRVVTIGPDTKELQKIQETQGAD 72
L I +P +A LP+ T +PA C A P R V + P+ ++ Q +
Sbjct 708 LPDIVVPIPDVSADLPVTT---VPAFCRGAPSAVSSP-RQVNVDPE----ERFQNSMKKL 759
Query 73 KVSSSVELCGGTHLSNAAQLE 93
K SS+ +CGG LSN L+
Sbjct 760 KAISSISICGGIALSNKDILD 780
> Hs7661636
Length=243
Score = 30.8 bits (68), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 14/51 (27%), Positives = 28/51 (54%), Gaps = 0/51 (0%)
Query 55 IGPDTKELQKIQETQGADKVSSSVELCGGTHLSNAAQLEDFVVISEESIAK 105
I PD + ++++E G + +S L GTH+ + +++ V+ E+ I K
Sbjct 142 IKPDMETYERLREKHGEEFFPTSNSLLHGTHVPSTEEIDRMVIDLEKQIEK 192
> At3g07690
Length=455
Score = 30.8 bits (68), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 37/126 (29%), Positives = 57/126 (45%), Gaps = 15/126 (11%)
Query 22 VNTAVLPLRTAME-LPALCAVFGEAYPDPVRVVTIGPDTKE---LQKIQETQGA--DKVS 75
V T +P R E L + V G+A DP+R+V +G + +QE G KVS
Sbjct 5 VETNSVPSRLVEERLDEIRRVMGKADDDPLRIVGVGAGAWGSVFIAMLQENYGKFRGKVS 64
Query 76 SSVELCGGTHLSNAAQLEDFVVIS--EESIAKGVRR------IVAVAGEAAISARSELRD 127
+ GG + A F VI+ EE + + +RR + A G+ + A L+D
Sbjct 65 VRIWRRGGRAIDKATAEHLFEVINSREELLRRLIRRCAYLKYVEARLGDRVLYADEILKD 124
Query 128 -VCLSL 132
CL++
Sbjct 125 GFCLNM 130
> At5g24100
Length=614
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 47/95 (49%), Gaps = 9/95 (9%)
Query 44 EAYPDPVRVVTIGPDTKELQKIQETQGADKVSSSVELCG-----GTHLSNAAQLEDFVVI 98
E PD +++ P KE+ K+ + + + + E+ G++L A LED ++
Sbjct 285 EPKPDKLKLAKKMPSEKEVSKLGKEKNIEDMEDKSEINKVMFFEGSNL--AFNLEDLLIA 342
Query 99 SEESIAKGVRRIV--AVAGEAAISARSELRDVCLS 131
S E + KGV + AV ++ + A L+D+ +S
Sbjct 343 SAEFLGKGVFGMTYKAVLEDSKVIAVKRLKDIVVS 377
> Hs14149744
Length=2069
Score = 29.6 bits (65), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 140 PAFPFFEKEVQQAKAFVESN--KMIPLLSRRRLLADLEN 176
PA FF +Q + F++ N K + ++RR +L+DLE+
Sbjct 1094 PATEFFGNTAEQTEPFLQQNRTKQVEGVTRRHVLSDLED 1132
> Hs7705947
Length=2047
Score = 29.3 bits (64), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 140 PAFPFFEKEVQQAKAFVESN--KMIPLLSRRRLLADLEN 176
PA FF +Q + F++ N K + ++RR +L+DLE+
Sbjct 1116 PATEFFGNTAEQTEPFLQQNRTKQVEGVTRRHVLSDLED 1154
Lambda K H
0.317 0.132 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5666887360
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40