bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
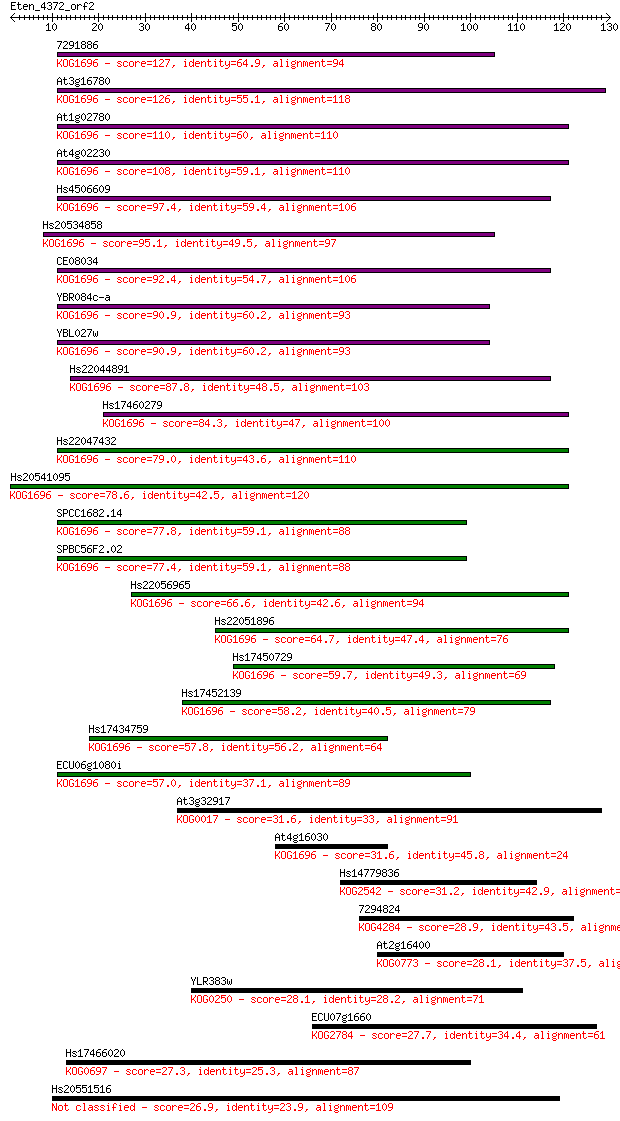
Query= Eten_4372_orf2
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
7291886 127 7e-30
At3g16780 126 9e-30
At1g02780 110 8e-25
At4g02230 108 3e-24
Hs4506609 97.4 6e-21
Hs20534858 95.1 3e-20
CE08034 92.4 2e-19
YBR084c-a 90.9 5e-19
YBL027w 90.9 5e-19
Hs22044891 87.8 5e-18
Hs17460279 84.3 5e-17
Hs22047432 79.0 2e-15
Hs20541095 78.6 3e-15
SPCC1682.14 77.8 5e-15
SPBC56F2.02 77.4 6e-15
Hs22056965 66.6 1e-11
Hs22051896 64.7 4e-11
Hs17450729 59.7 1e-09
Hs17452139 58.2 3e-09
Hs17434759 57.8 4e-09
ECU06g1080i 57.0 9e-09
At3g32917 31.6 0.34
At4g16030 31.6 0.39
Hs14779836 31.2 0.55
7294824 28.9 2.6
At2g16400 28.1 4.2
YLR383w 28.1 4.4
ECU07g1660 27.7 6.1
Hs17466020 27.3 7.4
Hs20551516 26.9 10.0
> 7291886
Length=203
Score = 127 bits (318), Expect = 7e-30, Method: Compositional matrix adjust.
Identities = 61/94 (64%), Positives = 75/94 (79%), Gaps = 0/94 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGT +AR+P K+LWM+RQRVLRRLLKKYRD+KKI H+YH LY++CKGN F
Sbjct 73 GRHCGFGKRKGTANARMPTKLLWMQRQRVLRRLLKKYRDSKKIDRHLYHDLYMKCKGNVF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKA 104
KNKRVL+E IH +K K + K L +Q +AR+QK
Sbjct 133 KNKRVLMEYIHKKKAEKQRSKMLADQAEARRQKV 166
> At3g16780
Length=209
Score = 126 bits (317), Expect = 9e-30, Method: Compositional matrix adjust.
Identities = 65/122 (53%), Positives = 84/122 (68%), Gaps = 4/122 (3%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGTR+ARLP K+LWMRR RVLRR L KYR++KKI HMYH +Y++ KGN F
Sbjct 73 GRHSGYGKRKGTREARLPTKILWMRRMRVLRRFLSKYRESKKIDRHMYHDMYMKVKGNVF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA----GFAAAK 126
KNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E + A G
Sbjct 133 KNKRVLMESIHKMKAEKAREKTLADQFEAKRIKNKASRERKFARREERLAQGPGGGETTT 192
Query 127 PA 128
PA
Sbjct 193 PA 194
> At1g02780
Length=214
Score = 110 bits (274), Expect = 8e-25, Method: Compositional matrix adjust.
Identities = 66/110 (60%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGTR+ARLP KVLWMRR RVLRRLLKKYR+ KKI HMYH +Y+R KGN F
Sbjct 73 GRHSGYGKRKGTREARLPTKVLWMRRMRVLRRLLKKYRETKKIDKHMYHDMYMRVKGNVF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 120
KNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E + A
Sbjct 133 KNKRVLMESIHKSKAEKAREKTLSDQFEAKRAKNKASRERKHARREERLA 182
> At4g02230
Length=208
Score = 108 bits (270), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 65/110 (59%), Positives = 82/110 (74%), Gaps = 0/110 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGTR+ARLP KVLWMRR RVLRRLLKKYR+ KKI HMYH +Y++ KGN F
Sbjct 73 GRHSGYGKRKGTREARLPTKVLWMRRMRVLRRLLKKYRETKKIDRHMYHDMYMKVKGNVF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 120
KNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E + A
Sbjct 133 KNKRVLMESIHKSKAEKAREKTLSDQFEAKRAKNKASRERKHARREERLA 182
> Hs4506609
Length=196
Score = 97.4 bits (241), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 63/106 (59%), Positives = 80/106 (75%), Gaps = 3/106 (2%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RHMGIGKRKGT +AR+P KV WMRR R+LRRLL++YR++KKI HMYH LY++ KGN F
Sbjct 73 GRHMGIGKRKGTANARMPEKVTWMRRMRILRRLLRRYRESKKIDRHMYHSLYLKVKGNVF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 116
KNKR+L+E IH K K ++K L +Q +AR+ K KE RK +EE
Sbjct 133 KNKRILMEHIHKLKADKARKKLLADQAEARRSKT---KEARKRREE 175
> Hs20534858
Length=376
Score = 95.1 bits (235), Expect = 3e-20, Method: Compositional matrix adjust.
Identities = 48/97 (49%), Positives = 68/97 (70%), Gaps = 2/97 (2%)
Query 8 CTPSRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKG 67
C RHMGIGK+K T +A++P KV WMR+ +L RLL++YR++KKI HMYH LY++ KG
Sbjct 282 CRKGRHMGIGKQKRTANAQIPEKVTWMRK--ILHRLLRRYRESKKIDGHMYHSLYLKVKG 339
Query 68 NQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKA 104
N FKNK +++E I+ K K +K L +Q +A + K
Sbjct 340 NVFKNKWIVMEHINKLKADKAHKKLLADQAEAPRSKT 376
> CE08034
Length=198
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 58/106 (54%), Positives = 74/106 (69%), Gaps = 3/106 (2%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKR+GT +AR+P K LW+RR RVLR LL++YRDAKK+ H+YH+LY+R KGN F
Sbjct 73 GRHTGYGKRRGTANARMPEKTLWIRRMRVLRNLLRRYRDAKKLDKHLYHELYLRAKGNNF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 116
KNK+ LIE I +K + K L +Q AR+ K KE RK +EE
Sbjct 133 KNKKNLIEYIFKKKTENKRAKQLADQAQARRDK---NKESRKRREE 175
> YBR084c-a
Length=189
Score = 90.9 bits (224), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 56/93 (60%), Positives = 68/93 (73%), Gaps = 0/93 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGTR+ARLP++V+W+RR RVLRRLL KYRDA KI H+YH LY KGN F
Sbjct 73 GRHSGYGKRKGTREARLPSQVVWIRRLRVLRRLLAKYRDAGKIDKHLYHVLYKESKGNAF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQK 103
K+KR L+E I K +EKAL E+ +AR+ K
Sbjct 133 KHKRALVEHIIQAKADAQREKALNEEAEARRLK 165
> YBL027w
Length=189
Score = 90.9 bits (224), Expect = 5e-19, Method: Compositional matrix adjust.
Identities = 56/93 (60%), Positives = 68/93 (73%), Gaps = 0/93 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGTR+ARLP++V+W+RR RVLRRLL KYRDA KI H+YH LY KGN F
Sbjct 73 GRHSGYGKRKGTREARLPSQVVWIRRLRVLRRLLAKYRDAGKIDKHLYHVLYKESKGNAF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQK 103
K+KR L+E I K +EKAL E+ +AR+ K
Sbjct 133 KHKRALVEHIIQAKADAQREKALNEEAEARRLK 165
> Hs22044891
Length=120
Score = 87.8 bits (216), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 50/103 (48%), Positives = 66/103 (64%), Gaps = 3/103 (2%)
Query 14 MGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNK 73
MGI KRKGT +A++P V WMRR R+L LL++Y ++KKI H YH LY++ KGN FKNK
Sbjct 1 MGISKRKGTANAQMPGNVTWMRRMRILCWLLRRYCESKKIDHHTYHSLYLKVKGNVFKNK 60
Query 74 RVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 116
+L+E I K K +K +Q AR+ K KE RK E+
Sbjct 61 WILMEHILKLKADKAHKKLQADQAKARRSKT---KEARKHHED 100
> Hs17460279
Length=170
Score = 84.3 bits (207), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 47/100 (47%), Positives = 65/100 (65%), Gaps = 3/100 (3%)
Query 21 GTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAI 80
GT +AR+P KV WM+R R+L LL++Y ++KKI HMYH LY++ +GN F NK +L+E
Sbjct 57 GTANARMPEKVTWMKRMRILHLLLRRYCESKKIDRHMYHSLYLKVQGNVFTNKPILMEHS 116
Query 81 HNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 120
H K K +K L +Q +AR+ K KE RK EE +A
Sbjct 117 HKLKADKAHKKLLADQAEARRPKT---KEARKRSEERLQA 153
> Hs22047432
Length=521
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/110 (43%), Positives = 65/110 (59%), Gaps = 16/110 (14%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RHMG GKRKGT +A +P KV +YRD+KKI HMYH LY++ K + F
Sbjct 73 GRHMGTGKRKGTANAPMPEKV-------------TRYRDSKKINHHMYHSLYLKVKESVF 119
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 120
K+K++L+E IH K K +K L +Q +A + K KE RK +EE +A
Sbjct 120 KDKQILMEHIHKLKADKAGKKLLADQAEACRPKT---KEARKQREECLQA 166
> Hs20541095
Length=240
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 51/124 (41%), Positives = 72/124 (58%), Gaps = 19/124 (15%)
Query 1 SSARPWPCTPS----RHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSH 56
S A+ W T + +HM IG+ V W+RR R+L RLL++Y ++KKI H
Sbjct 115 SQAQFWKNTLAHQKGKHMCIGQ------------VTWIRRTRILCRLLRRYHESKKIDCH 162
Query 57 MYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 116
MYH LY++ KGN FKNK++L+E I K K +K L +Q +AR+ K K+ RK EE
Sbjct 163 MYHSLYLQMKGNVFKNKQILMEYIRKLKAGKACKKLLADQAEARRSKT---KDARKHSEE 219
Query 117 NKKA 120
+A
Sbjct 220 RLQA 223
> SPCC1682.14
Length=193
Score = 77.8 bits (190), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 52/88 (59%), Positives = 64/88 (72%), Gaps = 0/88 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGT +AR+P+ V+WMRRQRVLRRLL+KYR++ KI H+YH LY+ KGN F
Sbjct 73 GRHTGYGKRKGTAEARMPSTVVWMRRQRVLRRLLRKYRESGKIDKHLYHTLYLEAKGNTF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVD 98
K+KR LIE I K + K +QEQ D
Sbjct 133 KHKRALIEHIQRAKAEANRTKLIQEQQD 160
> SPBC56F2.02
Length=193
Score = 77.4 bits (189), Expect = 6e-15, Method: Compositional matrix adjust.
Identities = 52/88 (59%), Positives = 64/88 (72%), Gaps = 0/88 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RH G GKRKGT +AR+P+ V+WMRRQRVLRRLL+KYR++ KI H+YH LY+ KGN F
Sbjct 73 GRHTGYGKRKGTAEARMPSAVVWMRRQRVLRRLLRKYRESGKIDKHLYHTLYLEAKGNTF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVD 98
K+KR LIE I K + K +QEQ D
Sbjct 133 KHKRALIEHIQRAKAEANRTKLIQEQQD 160
> Hs22056965
Length=108
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 40/94 (42%), Positives = 57/94 (60%), Gaps = 3/94 (3%)
Query 27 LPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNL 86
+ K WMRR R+L RLL++Y ++KK H+Y LY++ KGN FKNK++ E IH K
Sbjct 1 MSQKDTWMRRMRILCRLLRRYLESKKTDCHVYPSLYLKVKGNVFKNKQIFTEHIHKLKTD 60
Query 87 KVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 120
+ +K L +Q AR+ K KE+ K EE +A
Sbjct 61 EAHKKLLADQAVARRSKT---KEEHKHHEERLQA 91
> Hs22051896
Length=145
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 36/76 (47%), Positives = 52/76 (68%), Gaps = 3/76 (3%)
Query 45 KKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKA 104
++Y ++KK+ HMYH LY++ KGN FKNKR+L+E IHN K K +K L +Q + R+ K
Sbjct 63 RRYHESKKVDRHMYHSLYLKVKGNVFKNKRILMEHIHNLKADKACKKLLADQAETRRSKI 122
Query 105 AAQKEKRKLKEENKKA 120
KE RK +EE+ +A
Sbjct 123 ---KEARKHREEHLQA 135
> Hs17450729
Length=147
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/69 (49%), Positives = 46/69 (66%), Gaps = 3/69 (4%)
Query 49 DAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQK 108
++KKI HMYH LY++ KGN FK+KR+L E H K K ++K L +Q +AR K K
Sbjct 62 ESKKIDRHMYHSLYLKLKGNVFKHKRILTEHSHKLKADKARKKPLADQAEARGSKT---K 118
Query 109 EKRKLKEEN 117
E RKL+EE+
Sbjct 119 EARKLREEH 127
> Hs17452139
Length=118
Score = 58.2 bits (139), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/79 (40%), Positives = 47/79 (59%), Gaps = 3/79 (3%)
Query 38 RVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQV 97
R+L L +Y ++KK HMYH LY++ +GN F NK++L+E H K K +K L +Q
Sbjct 2 RILHWLFGRYHESKKTDHHMYHSLYLKVQGNMFTNKQILMEHNHKLKADKAHKKFLADQA 61
Query 98 DARKQKAAAQKEKRKLKEE 116
+AR K + KL+EE
Sbjct 62 EARSSKT---NKASKLREE 77
> Hs17434759
Length=138
Score = 57.8 bits (138), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/64 (56%), Positives = 49/64 (76%), Gaps = 0/64 (0%)
Query 18 KRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLI 77
K+KGT +AR+P K+ WMRR R+LR LL++Y ++KKI +MYH LY++ KGN FKNK +L
Sbjct 72 KQKGTANARMPEKITWMRRMRILRLLLRRYCESKKIDCYMYHSLYLKVKGNVFKNKWILR 131
Query 78 EAIH 81
E H
Sbjct 132 EHSH 135
> ECU06g1080i
Length=171
Score = 57.0 bits (136), Expect = 9e-09, Method: Compositional matrix adjust.
Identities = 33/89 (37%), Positives = 51/89 (57%), Gaps = 0/89 (0%)
Query 11 SRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQF 70
RHMG GKR GT +AR+P K +WM++ R +R +LK+ R +I + Y + KG+ F
Sbjct 73 GRHMGTGKRFGTANARMPQKKIWMKKIRAMRAMLKEMRANGEITKEDFRVFYKQAKGHLF 132
Query 71 KNKRVLIEAIHNEKNLKVKEKALQEQVDA 99
K++ + + I K + + K L EQ A
Sbjct 133 KHRFHMKDCIIKRKADEQRAKELAEQAKA 161
> At3g32917
Length=757
Score = 31.6 bits (70), Expect = 0.34, Method: Composition-based stats.
Identities = 30/101 (29%), Positives = 46/101 (45%), Gaps = 21/101 (20%)
Query 37 QRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIE-AIHNEKNLK-------- 87
Q +RR L+ R L VRC+ +Q+ K L+E A+ E++L+
Sbjct 251 QAQMRRFLRGLRP----------DLRVRCRVSQYATKAALVETAVKVEEDLQRQVVAVSR 300
Query 88 -VKEKALQEQVDARKQKAAAQKEKRKLKEENKKAGFAAAKP 127
V+ K Q+QV K AQ +KRK + +AG +P
Sbjct 301 AVQPKKTQQQVAPSKGNKPAQGQKRKW-DHPSRAGQGHLRP 340
> At4g16030
Length=101
Score = 31.6 bits (70), Expect = 0.39, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 19/24 (79%), Gaps = 0/24 (0%)
Query 58 YHKLYVRCKGNQFKNKRVLIEAIH 81
YH ++++ KG +KNK VL+E++H
Sbjct 37 YHDMFMKVKGKVYKNKCVLMESMH 60
> Hs14779836
Length=509
Score = 31.2 bits (69), Expect = 0.55, Method: Composition-based stats.
Identities = 18/42 (42%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 72 NKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKL 113
N R L EA+ E L++ E L E+ DA Q+ QK +RKL
Sbjct 228 NLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKL 269
> 7294824
Length=1028
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 23/46 (50%), Gaps = 14/46 (30%)
Query 76 LIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKAG 121
L E+ HN KNL+VKEK + Q KLKE N KAG
Sbjct 488 LPESSHNNKNLRVKEKEIVRQ--------------GKLKETNSKAG 519
> At2g16400
Length=482
Score = 28.1 bits (61), Expect = 4.2, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 80 IHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKK 119
IHN K LK ++ L E V+ +K Q E K+ E +K
Sbjct 115 IHNSKYLKAAQELLDETVNVKKALKQFQPEGDKINEVKEK 154
> YLR383w
Length=1114
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 20/74 (27%), Positives = 38/74 (51%), Gaps = 3/74 (4%)
Query 40 LRRLLKKYRDAKKIVSHMYHKLYVRCKGNQFKNKRVLIEAIHNE---KNLKVKEKALQEQ 96
L+ L +Y DAKK++ + + + + K + I+ HN KNL+ + +Q++
Sbjct 277 LKSLKAEYEDAKKLLRELNQTSDLNERKMLLQAKSLWIDVAHNTDACKNLENEISGIQQK 336
Query 97 VDARKQKAAAQKEK 110
VD +K ++EK
Sbjct 337 VDEVTEKIRNRQEK 350
> ECU07g1660
Length=475
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 21/66 (31%), Positives = 32/66 (48%), Gaps = 5/66 (7%)
Query 66 KGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKE-----KRKLKEENKKA 120
K N F+NK + E +N++ E ++ V + K+ A+ +E KRKL KK
Sbjct 93 KMNAFRNKWIRREGNRVFRNVESIEDTVRNMVISMKRGTASAEEVEALRKRKLVSRRKKI 152
Query 121 GFAAAK 126
F A K
Sbjct 153 VFIATK 158
> Hs17466020
Length=228
Score = 27.3 bits (59), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 41/88 (46%), Gaps = 2/88 (2%)
Query 13 HMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCK-GNQFK 71
H GI ++A L K L R + ++++ + D +V + ++ R G +
Sbjct 140 HKGISPEAAKKEAEL-GKYLECRVEEIIKKQGEGVPDLVHVVYTLANENIPRLPPGGELA 198
Query 72 NKRVLIEAIHNEKNLKVKEKALQEQVDA 99
NK+ +IEAI+ +NL + + DA
Sbjct 199 NKQNVIEAIYKRRNLYKNDDSDSTSTDA 226
> Hs20551516
Length=1723
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 26/109 (23%), Positives = 39/109 (35%), Gaps = 0/109 (0%)
Query 10 PSRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIVSHMYHKLYVRCKGNQ 69
P H G+G+ T LPAK + + L + S HK K +
Sbjct 216 PRNHFGVGRSTVTTKVTLPAKPKHVELNLKTPKNLDSLGNEHNPFSQPVHKGNTATKISL 275
Query 70 FKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENK 118
F+NKR H + + A + R + A+K K + E K
Sbjct 276 FENKRTNSSPRHTDIRGQRNTPASSKTFVGRAKLNLAKKAKEMEQPEKK 324
Lambda K H
0.318 0.129 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40