bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_4384_orf1
Length=78
Score E
Sequences producing significant alignments: (Bits) Value
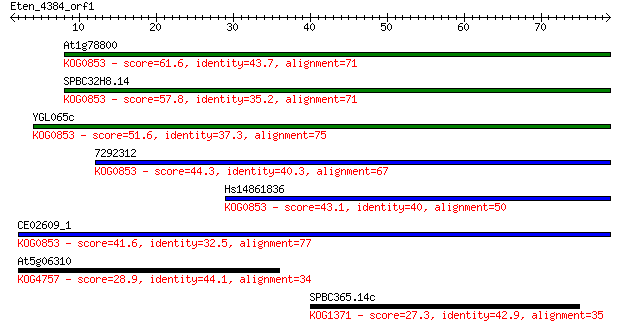
At1g78800 61.6 3e-10
SPBC32H8.14 57.8 4e-09
YGL065c 51.6 3e-07
7292312 44.3 6e-05
Hs14861836 43.1 1e-04
CE02609_1 41.6 4e-04
At5g06310 28.9 2.2
SPBC365.14c 27.3 7.5
> At1g78800
Length=405
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/72 (43%), Positives = 43/72 (59%), Gaps = 2/72 (2%)
Query 8 LNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV 67
+ V+GSFLP I C+ LR L++ L ++ G +D+V DQV+VV PLL+L
Sbjct 62 FQVTVYGSFLPRHIFYRLHAVCAYLRCLFVAL-CVLLGWSSFDVVLADQVSVVVPLLKLK 120
Query 68 -GKKVIFYGHFP 78
KV+FY HFP
Sbjct 121 RSSKVVFYCHFP 132
> SPBC32H8.14
Length=511
Score = 57.8 bits (138), Expect = 4e-09, Method: Composition-based stats.
Identities = 25/71 (35%), Positives = 43/71 (60%), Gaps = 1/71 (1%)
Query 8 LNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV 67
+ + V+G +LP SI +FCS LR +YL + ++T ++ +D + DQ++ P L L
Sbjct 70 IKVKVYGDWLPSSIFGRLSIFCSSLRQVYLTMI-LLTNYMHFDAIIVDQLSTCVPFLLLA 128
Query 68 GKKVIFYGHFP 78
+ ++FY HFP
Sbjct 129 SQMILFYCHFP 139
> YGL065c
Length=503
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 2/77 (2%)
Query 4 KTSRLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHV-GYDLVFNDQVAVVNP 62
K +L + V+G FLP + L F + + +R LYLV+ I+ V Y L+ DQ++ P
Sbjct 56 KNGQLKVEVYGDFLPTNFLGRFFIVFATIRQLYLVIQLILQKKVNAYQLIIIDQLSTCIP 115
Query 63 LLRLVGK-KVIFYGHFP 78
LL + ++FY HFP
Sbjct 116 LLHIFSSATLMFYCHFP 132
> 7292312
Length=424
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 27/71 (38%), Positives = 36/71 (50%), Gaps = 4/71 (5%)
Query 12 VFGSFLPHSILRHFRVFCSVLRMLYLVLTA--IMTGHVGYDLVFNDQVAVVNPLLRLVG- 68
V G +LP + F C+ LRMLY + A M D+V D ++V P+LR
Sbjct 59 VVGDWLPRGLFGRFYAICAYLRMLYAAIYASFFMPQREQVDVVVCDLISVCIPVLRFAPH 118
Query 69 -KKVIFYGHFP 78
KV+FY HFP
Sbjct 119 RPKVLFYCHFP 129
> Hs14861836
Length=416
Score = 43.1 bits (100), Expect = 1e-04, Method: Composition-based stats.
Identities = 20/52 (38%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 29 CSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVNPLLRLV--GKKVIFYGHFP 78
C+ +RM++L L + +D+V DQV+ P+ RL KK++FY HFP
Sbjct 89 CAYVRMVFLALYVLFLADEEFDVVVCDQVSACIPVFRLARRRKKILFYCHFP 140
> CE02609_1
Length=384
Score = 41.6 bits (96), Expect = 4e-04, Method: Composition-based stats.
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 2/78 (2%)
Query 2 LQKTSRLNIVVFGSFLPHSILRHFRVFCSVLRMLYLVLTAIMTGHVGYDLVFNDQVAVVN 61
Q+T L+I ++P SI C+ L+M+ L I+ H D++ +D V+
Sbjct 47 FQETLDLDICTVVPWIPRSIFGKGHALCAYLKMIIAALY-IVIYHKDTDVILSDSVSASQ 105
Query 62 PLLRLVGK-KVIFYGHFP 78
+LR K K++FY HFP
Sbjct 106 FVLRHFSKAKLVFYCHFP 123
> At5g06310
Length=463
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 2 LQKTSRLNIVVFGSFLPHSILRHFRVFCSVLRML 35
L K+ RL + V LP +LR F F SVLR++
Sbjct 200 LVKSERLPLCVEPEMLPTYMLRKFPTFGSVLRII 233
> SPBC365.14c
Length=355
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 23/40 (57%), Gaps = 5/40 (12%)
Query 40 TAIMTGHVGYDLVF-----NDQVAVVNPLLRLVGKKVIFY 74
T ++ GYD+V N +V V+ + +L GKKVIF+
Sbjct 22 TCVVLLEKGYDVVIVDNLCNSRVEAVHRIEKLTGKKVIFH 61
Lambda K H
0.336 0.149 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171925608
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40